- Home
- News
- Spotlight on Science
- How chemicals can...
How chemicals can combine to form deadly cocktails from harmless components
01-10-2015
We are constantly exposed to external compounds, so-called xenobiotics, in our environment. Most chemicals in use today in food and drugs have passed stringent safety tests and are not individually harmful to human health. However, combinations of these chemicals can have a very different effect to the individual compound, with profound implications for human health. Many of these chemicals target nuclear receptors, proteins that control gene expression networks in response to signalling molecules. Crystal structures of a human nuclear receptor, the pregnane X receptor, have been solved in complex with oestrogen and a pesticide showing the synergistic effect that simultaneous binding has on the protein compared to the individual compounds. The results have the potential to change current policies in risk assessment for chemicals and could lead to improved chemicals in the future.
Human nuclear receptors control the expression of genes in response to the binding of small signalling molecules in the body. Many compounds that are in the environment are classed as endocrine-disrupting chemicals as they are thought to directly bind nuclear receptors and change gene expression patterns, leading to harmful physical effects. While many of these compounds are harmless individually, the body is constantly exposed to mixtures of these compounds in the environment. It has long been known that combinations of these chemicals can have additive, antagonistic or synergistic effects, but there has been no understanding of the molecular basis for these effects.
The teams of Patrick Balaguer and William Bourguet of INSERM, CNRS and the University of Montpellier used extensive screening methods and biophysical characterisation to identify two endocrine-disrupting chemicals that act synergistically on a human nuclear receptor, the pregnane X receptor (PXR). The two compounds identified are a pesticide, trans-nonachlor, and the active ingredient of the contraceptive pill, the oestrogen derivative 17α-ethinylestradiol; two compounds with very different uses that are commonly found in our environment. By solving crystal structures of PXR bound to the two chemicals individually and in combination, the teams have been able to propose a molecular mechanism for the observed synergistic effect between the molecules.
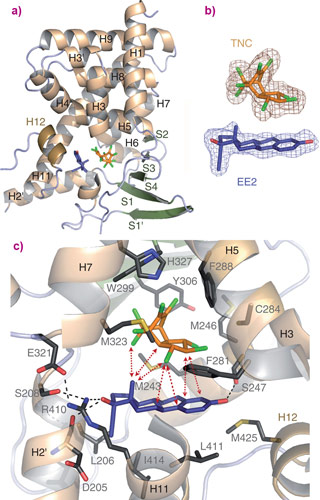
The crystal structures show that the two chemicals bind in a different manner together than individually forming a ‘supramolecular’ ligand causing a sustained activation of the receptor (Figure 1). The binding pocket of PXR is relatively large as a diverse range of molecules need to be accommodated. A large number of contacts between the two different compounds allow the binding pocket to be tightly held in the active conformation, leading to the observed physiological effects.
Many conditions had to be tested in order to obtain crystals of the PXR in complex with various combinations of xenobiotics. This means that the diffraction properties of hundreds of crystals needed to be examined in a synchrotron X-ray beam in order to select those that diffract to a sufficient resolution to allow proper visualisation of drug binding. This type of sample evaluation is only possible at the ESRF Structural Biology beamlines due to their high level of automation of beam alignment and sample handling. The final data sets were collected at the microfocus beamline ID23-2.
Article written by M.W. Bowler (EMBL, Grenoble).
Principal publication and authors
Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds, V. Delfosse (a,b,c), B. Dendele (c,d,e,f), T. Huet (a,b,c), M. Grimaldi (c,d,e,f), A. Boulahtouf (c,d,e,f), S. Gerbal-Chaloin (c,g), B. Beucher (a,h,i), D. Roecklin (j),C. Muller (j), R. Rahmani (k), V. Cavaille (c,d,e,f), M. Daujat-Chavanieu (c,g,l), V. Vivat (j),J-M. Pascussi (c,i,j), P. Balaguer (c,d,e,f) & W. Bourguet (a,b,c), Nature Communications, 6, 8089 (2015).
(a) Inserm U1054, Montpellier (France)
(b) CNRS UMR5048, Centre de Biochimie Structurale, Montpellier (France)
(c) Universite de Montpellier (France)
(d) IRCM, Institut de Recherche en Cancerologie de Montpellier (France)
(e) Inserm, U1194, Montpellier (France)
(f) ICM, Institut regional du Cancer de Montpellier (France)
(g) Inserm U1040, Montpellier (France)
(h) Inserm U661, Montpellier (France)
(i) CNRS UMR5203, Institut de Genomique Fonctionnelle, Montpellier (France)
(j) NovAliX, Illkirch (France)
(k) INRA UMR 1331,TOXALIM, Sophia-Antipolis (France)
(l) CHU de Montpellier, Institut de Recherche en Biotherapie (France)
Top image: Trans-nonachlor, a pesticide, and 17α-ethinylestradiol, the active ingredient of the contraceptive pill, combine within a human nuclear receptor forming a ligand with enhanced activity (Image credit: Delfosse, V. et al.).