- Home
- News
- Spotlight on Science
- Coiled-coil protein-origami...
Coiled-coil protein-origami cages self-assemble in cells
18-12-2017
Protein cages based on coiled coil units can self-assemble in bacteria and mammalian cells. A tetrahedron, rectangular pyramid and trigonal prism cages of 5 to 10 nm diameters were designed. The prism, composed of more than 700 amino acid residues, represents the largest single-chain designed protein. Potential applications include drug delivery, vaccines, enzyme encapsulation, smart biomaterials, biosensors and molecular machines.
Proteins are nature’s nanomachines performing key processes necessary to sustain life, such as catalysis of chemical reactions, transportation of oxygen in our blood and molecular recognition. Proteins also provide structural support, for example in the form of microtubules, actin filaments, intermediary filaments and collagen fibres. Proteins are built as a linear chain of amino acids, which encodes information about the 3D structure. Before proteins can do their jobs, they must first fold into the correct 3D structure. Understanding the relationship between the sequence, 3D structure and function of proteins is one of the key challenges of protein research. The problem is not only of academic interest but also of great technological importance as designed proteins could be used to build new nanomaterials, transport vessels and therapeutic agents such as improved vaccines, cancer therapeutics and flu inhibitors. However, designing novel proteins is far from trivial. Twenty different amino acids appear in proteins, making the number of possible amino acid sequences almost infinite. For peptides with 8 amino acids, there are four times as many possible sequences as there are people on earth. For proteins of length 64, there are more possible sequences than the estimated number of all atoms in the known universe!
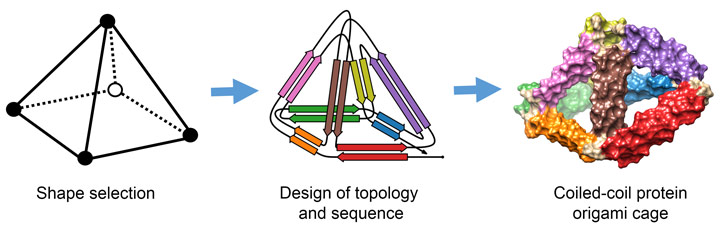
Scientist from the Department of Synthetic Biology and Immunology at the National Institute of Chemistry in Ljubljana, Slovenia have found an alternative solution to the protein-folding puzzle. They have invented a method to design protein cages of any selected shape that can self-assemble in living organisms. Their approach was based on previous work that included the design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments [1]. The method was named “coiled-coil protein origami” (CCPO), since it was inspired by DNA origami, a technique that involves folding a long DNA strand into the desired shape by multiple short complementary DNA staples [2].
The key insight of CCPO was to use coiled-coils as building blocks. Coiled-coils are natural protein binding motifs that can be designed so that only the specific pairs form in solution. In this way, a toolbox has been created of coiled-coil building modules with simple and modular binding rules.
To construct larger structures, the coiled-coils have to be connected together in a longer chain. The scientists, assisted by the mathematician Tomaž Pisanski, solved the folding problem in a way that can be likened to the construction of a pyramid from a single piece of rope (Figure 1). They identified a path for the protein chain that passes over each side of the pyramid exactly twice. This path determines the order in which the halves of the coiled-coils have to be connected along the chain. Instructions for this strategy are available in an open-source program CoCoPOD [3].
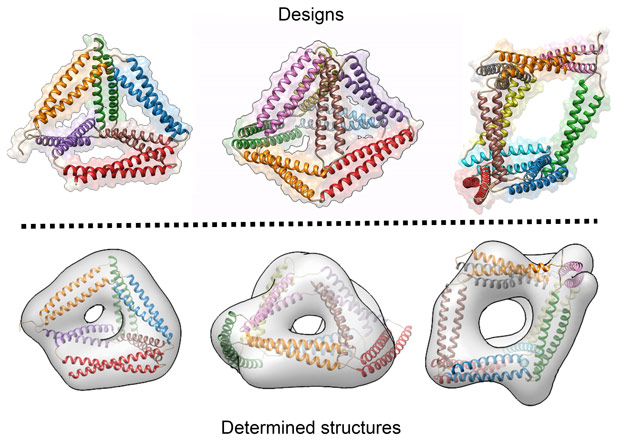
Over 20 variants of CCPO cages have been designed and characterised. Several tetrahedra, two square pyramids and a triangular prism were designed and produced in bacteria. The triangular prism at more than 700 amino-acids represents the largest de novo designed single chain protein so far. The cages correctly self-assemble in bacteria, mammalian cells and live mice, without causing unwanted side effects. The structures of the cages were confirmed using a variety of experimental techniques including small-angle X-ray scattering at beamline BM29 and electron microscopy and they agree well with the designed scaffolds (Figure 2).
 |
|
Figure 2. Designed and determined structures. The designed structures (top) match very well with structures determined from electron microscopy and SAXS modelling (bottom). |
The research was highly interdisciplinary, employing a wide range of techniques, from molecular and structural biology to immunology and even mathematics.
A multitude of potential applications have been envisioned: active substances could be packed inside the cages, leading to novel cargo delivery vehicles. For example, toxins could be encapsulated and the cages used to deliver the toxins only to cancer cells. Additionally, more efficient vaccines could be created, by placing antigens at the vertices of the cages. Future projects include the creation of cages that can open and close in response to external stimuli, such as light or pH, leading to the development of smart nanomaterials and biosensors and perhaps novel molecular machines.
Principal publication and authors
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo, A. Ljubetič (a) , F. Lapenta (a,b), H. Gradišar (a,c), I. Drobnak (a), J. Aupič (a,d), Ž. Strmšek (a), D. Lainšček (a), I. Hafner-Bratkovič (a,c), A. Majerle (a), N. Krivec (a), M. Benčina (a), T. Pisanski (e), T. Ćirković Veličković (f), A. Round (g,h), J. María Carazo (i), R. Melero (i) and R. Jerala (a,c), Nat Biotechnol. 35, 1094-1101 (2017); doi: 10.1038/nbt.3994.
(a) Department of Synthetic Biology and Immunology, National Institute of Chemistry, Ljubljana (Slovenia)
(b) Graduate School of Biomedicine, University of Ljubljana (Slovenia)
(c) EN-FIST Centre of Excellence, Ljubljana (Slovenia)
(d) Doctoral Study Programme in Chemical Sciences, University of Ljubljana (Slovenia)
(e) FAMINT, University of Primorska, Koper and Institute of Mathematics and Physics, Ljubljana (Slovenia)
(f) Center of Excellence for Molecular Food Sciences, Faculty of Chemistry, University of Belgrade (Serbia)
(g) ESRF
(h) School of Chemical and Physical Sciences, Keele Univerity (UK)
(i) Centro Nacional de Biotecnología (CNB-CSIC), Madrid (Spain)
References
[1] H. Gradišar et al., Nature Chemical Biology 9, 362–366 (2013); doi: 10.1038/nchembio.1248.
[2] P.W.K. Rothemund, Nature 440, 297–302 (2006); doi: 10.1038/nature04586.
[3] CoCoPOD: https://github.com/NIC-SBI/CC_protein_origami.
Acknowledgements
This project was funded by the Slovenian Research Agency, ERANET project Bioorigami, MSCA project Tollerant and a grant from ICGEB.