SAXSy DNA origami
Small-angle X-ray scattering (SAXS) was used to quantitatively determine the conformational changes of a DNA origami switch device as a function of ionic strength in free solution.
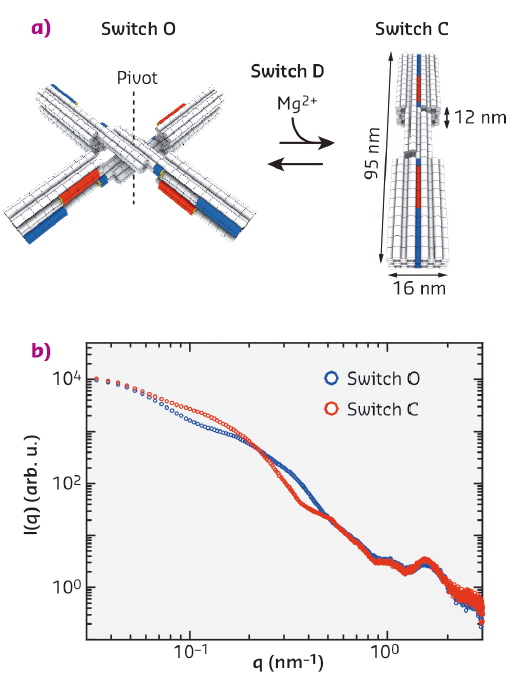
Molecular self-assembly is a ubiquitous process in nature describing the organisation of molecules into complex structural arrangements of high spatial accuracy such as protein-based enzymes and molecular machines. To create biomimetic objects that can adopt specific conformations and carry out functions at the molecular scale ranging from nanometres to micrometres, such as in transport, signal transduction, and molecular circuitry, DNA has emerged as an ideal building block due to its chemical addressability and predictable Watson−Crick base-pairing. In this context, the development of the DNA origami technique introduced by Paul Rothemund [1] ten years ago is a significant landmark of structural DNA nanotechnology. Here, a several kilobase long circular single-stranded 'scaffold strand' is folded with the assistance of hundreds of short single-stranded 'staple strands' resulting in predefined, precisely controlled shapes reaching over 100 nm in size and molecular weights of several MDa [2]. Examples include both static structures in two- or three-dimensions and dynamic devices that can undergo controlled conformational changes. Recently, Gerling et al. [3] established a framework based on shape-complementary recognition and nucleobase stacking interactions to construct a robust and controllable DNA "switch" device (Figure 80a). The switch structure can change between an open and closed state upon varying solution conditions such as salt concentration and temperature.
 |
|
Fig. 80: Schematics of the DNA origami switch devices used in this study and scattering profiles of the static open ("O") and closed ("C") switch samples. |
Structural characterisation of DNA origami structures has predominantly relied on atomic force microscopy (AFM) imaging and negative-stain transmission electron microscopy (TEM), requiring immobilisation of the samples on a surface and making it challenging to detect conformational changes upon variation in solution conditions. This study demonstrates that small-angle X-ray scattering (SAXS) can quantitatively determine the conformational changes of a DNA origami switch device as a function of ionic strength in free solution. In addition, our work highlights the ability of SAXS to evaluate structural models against solution-based data, even for very large and complex DNA objects, which constitutes a promising and complementary approach to surface-based methods.
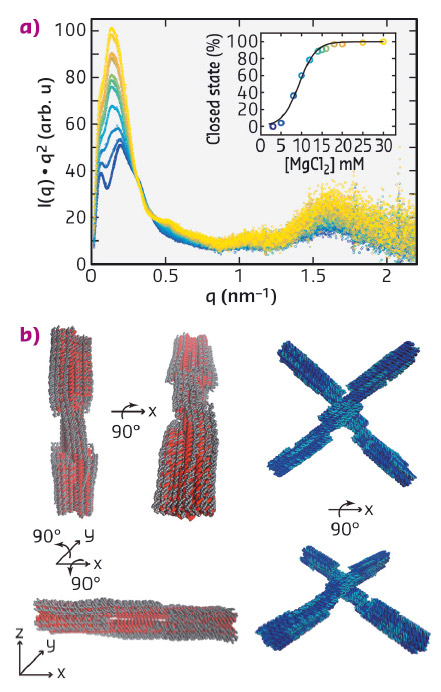
We performed solution SAXS measurements at beamline BM29 on static switch samples, which are permanently locked in the open (switch O) and closed conformation (switch C), respectively, and on a dynamic switch variant (switch D), which changes its conformation from the open to the closed state upon addition of magnesium ions (Figure 80a). For the static samples measured at concentrations of 25 - 100 nM, we obtained scattering profiles with a dynamic range of ≥ 4 orders of magnitude in intensity with features identifiable up to q ≈ 3 nm−1 (Figure 80b). To characterise the conformational transition from the open to the closed state of the dynamic switch variant upon the addition of MgCl2, we performed SAXS experiments at various MgCl2 concentrations, ranging from 3 to 30 mM (Figure 81a). The resulting scattering profiles could be described by a two-state fit, where the scattering data at each MgCl2 concentration is a linear superposition of the scattering profiles of the static open and closed conformations, allowing for the determination of the populations at each salt concentration. The salt-dependence of the fraction closed could be fitted by a thermodynamic model that assumes a linear dependence of the free energy ΔG on the ion concentration c (Figure 81b).
 |
|
Fig. 81: a) Kratky representation of the scattering profiles of switch D samples for MgCl2 concentrations ranging from 3 mM (dark blue, bottom) to 30 mM (light yellow, top). Inset: fraction of closed switch particles determined from SAXS and fit of a two-state model (black line). b) Structural models derived from the normal mode refinement. Initial models are shown in red and dark blue, refined models in grey and cyan. |
In addition, we compared our experimental data to scattering profiles of the static open and closed switch samples predicted from computer generated, idealised three-dimensional atomistic models, revealing systematic deviations between the experimental and theoretical profiles. We employed a normal mode based elastic network model approach to refine the idealised atomistic models against the experimental SAXS data. The results of the refinement suggest that the DNA helices are deformed from the idealised geometries, consistent with electrostatic repulsion, and reveal deviations from the idealised design geometries that are otherwise difficult to resolve. Our results establish SAXS as a powerful tool to investigate conformational changes and solution structures of DNA origami and we anticipate our methodology to be broadly applicable to increasingly complex DNA and RNA devices.
Principal publication and authors
Conformational changes and flexibility of DNA devices observed by small-angle X-ray scattering, L.K. Bruetzel (a), T. Gerling (b), S.M. Sedlak (a), P.U. Walker (a), W. Zheng (c), H. Dietz (b) and J. Lipfert (a), Nano Lett. 16, 4871−4879 (2016); doi: 10.1021/acs.nanolett.6b01338.
(a) Department of Physics, Nanosystems Initiative Munich, and Center for Nanoscience, LMU Munich (Germany)
(b) Department of Physics, Walter Schottky Institute, TUM Munich (Germany)
(c) Physics Department, State University of New York at Buffalo (USA)
References
[1] P.W.K. Rothemund, Nature, 440, 297–302, (2006).
[2] H. Dietz et al., Science 325, 725–730 (2009).
[3] T. Gerling et al., Science 347, 1446–1452 (2015).



