- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2015
- Structural biology
- Mechanistic clues from the structure of mitochondrial complex I
Mechanistic clues from the structure of mitochondrial complex I
The crystal structure of mitochondrial complex I, a central player in cellular energy conversion, provided important insights into the mechanism and regulation of this giant membrane protein.
Complex I (proton pumping NADH:ubiquinone oxidoreductase) [1] is the first component of the respiratory chain in the mitochondrial inner membrane. Mitochondria gain energy by oxidising hydrogen extracted from nutrients, converting it into a proton gradient that is then used to synthesise adenosine tri-phosphate (ATP), the universal energy currency of the cell. Complex I couples the transfer of two electrons from NADH (reduced nicotinamide adenine dinucleotide) to ubiquinone with the pumping of four protons across the inner mitochondrial membrane. Defects in human complex I are the most frequent cause of inherited mitochondrial disorders and are implicated in numerous (neuro)degenerative diseases and ageing. The mitochondrial, but not the bacterial complex, is regulated by the so-called active/deactive (A/D) transition. Arresting the enzyme in the deactive conformation by modifying a specific cysteine residue [2] protects the heart from reperfusion injury by preventing complex I from producing reactive oxygen species.
Mitochondrial complex I is a very large membrane integral multiprotein assembly of more than 40 different subunits. 14 central subunits harbour the catalytic function and are conserved from bacteria to human. The functions of most of the remaining accessory subunits are still elusive.
We crystallised intact mitochondrial complex I (~1 MDa, 41 subunits) from the yeast Yarrowia lipolytica. Using initial phase information from TaBr derivatives, we had previously obtained a first electron density map at 6.3 Å resolution with data measured at several beamlines at the ESRF and the Swiss Light Source [3]. Since then, markedly improved resolution was achieved through extensive optimisation of cryo-conditions, including rapid crystal cooling in liquid propane, and of data collection strategies that took advantage of the excellent beam quality at ID29 and ID23-1 coupled with Pilatus detectors. Using the microfocus beamline ID23-2, we could probe very large complex I crystals and conclude that they were largely homogeneous. This allowed the use of a larger beam size to illuminate a larger crystal volume yielding better datasets. Ultimately, datasets from the best crystal - diffracting X-rays anisotropically to 3.6 x 3.9 x 3.9 Å resolution - were collected at 5 K using helium cooling on beamline PXI at SLS. More phase information was obtained from additional derivatives, from the anomalous signal of the iron contained in the complex, and from selenomethionine labelled complex I.
Our current complex I structure comprises the 14 central subunits and the largest accessory subunit, with about half of the residues with side chains and the others as polyalanine chains, and additional polyalanine traces of accessory subunits. Overall, complex I is L-shaped consisting of a peripheral arm pointing into the mitochondrial matrix and a membrane arm integrated in the inner mitochondrial membrane (Figure 117). NADH oxidation occurs in the peripheral arm delivering electrons via a series of seven iron-sulfur clusters to the ubiquinone reduction pocket, which we could clearly identify using anomalous signals of a brominated substrate inhibitor analogue. From a narrow opening in the membrane domain, the pocket reaches about 30 Å into the peripheral arm and connects to a long ‘central axis’ of acidic and basic amino acid residues resembling an electric wire running through the middle of the entire membrane arm thereby connecting four putative proton pathways crossing the membrane. This remarkable arrangement spatially separates electron transfer and proton pumping covering a distance of more than 300 Å. Since the quinone-binding pocket connects these two functional branches, it seems likely that the redox chemistry of ubiquinone plays a pivotal role in transmitting energy into the membrane arm via the ‘central axis’ to drive proton translocation [1].
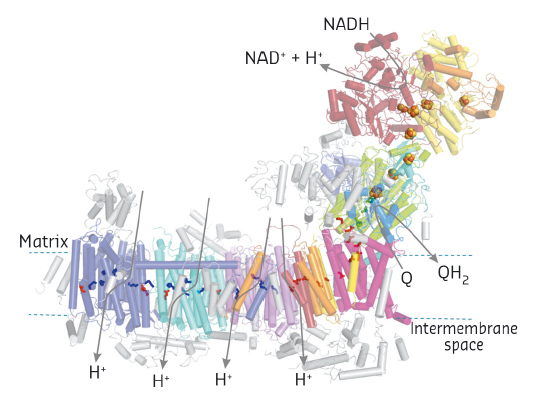 |
|
Fig. 117: Structural overview and functional topology of mitochondrial complex I. Accessory subunits in grey. Iron-sulfur clusters in space fill and selected of the central axis residues (see text) in stick representation. |
Since we crystallised the deactive form, our structure provided important clues to the conformational changes associated with complex I regulation. Relocation of the ubiquinone-binding site seemed to render the enzyme deactive by moving the pocket away from the iron-sulfur clusters. Intriguingly, this implicates a defined rearrangement of surrounding structural elements including the loop carrying the cysteine controlling the A/D transition and a loop of the first membrane integral subunit comprising the start of the ‘central axis’. Thereby our X-ray data provide a structural basis, for the active/deactive transition and for a previously proposed hypothetical energy conversion mechanism [4]. According to this mechanism, stabilisation of negatively charged ubiquinone intermediates provides the energy for specific conformational changes that are transmitted into the membrane arm to drive proton pumping of complex I.
Principal publication and authors
Mechanistic insight from the crystal structure of mitochondrial complex I, V. Zickermann (a,b), C. Wirth (c), H. Nasiri (d,e), K. Siegmund (a), H. Schwalbe (b,e), C. Hunte (c) and U. Brandt (b,f), Science 347, 44-49 (2015); doi: 10.1126/science.1259859.
(a) Structural Bioenergetics Group, Institute of Biochemistry II, Medical School, Goethe-University, Frankfurt am Main (Germany)
(b) Cluster of Excellence Frankfurt “Macromolecular Complexes”, Goethe-University, Frankfurt am Main (Germany)
(c) Institute of Biochemistry and Molecular Biology, BIOSS Centre for Biological Signalling Studies, University of Freiburg (Germany)
(d) Department of Chemistry, University of Cambridge (UK)
(e) Institute of Organic Chemistry and Chemical Biology, Goethe-University, Frankfurt am Main (Germany)
(f) Nijmegen Center for Mitochondrial Disorders, Radboudumc, Nijmegen (Netherlands)
References
[1] U. Brandt, Annu. Rev. Biochem. 75, 69-92 (2006).
[2] A. Galkin et al., J. Biol. Chem. 283, 20907-20913 (2008).
[3] C. Hunte et al., Science 329, 448-451 (2010).
[4] U. Brandt, Biochim. Biophys. Acta 1807, 1364-1369 (2011).



