EWOKS brings automated and reproducible data processing to ESRF beamlines
The ESRF has developed a major new tool to automate experiment and data processing, aiming to increase experimental throughput. The ESRF Workflow System (EWOKS) supports fully automated workflows on the beamlines but also brings the power of these workflows to users for local use. EWOKS thereby aims to make the processing of data more efficient, accessible and reproducible.
Share
The ESRF’S Extremely Brilliant Source (ESRF-EBS), launched in August 2020, has resulted in an increased demand in experiments demanding higher resolution and faster data collection. Therefore, the automation of beamlines and data processing is becoming increasingly crucial in order to speed up experiment throughput, data collection and processing (e.g., on-the-fly feedback on the quality of the collected data, data conversion or reduction, etc.). Automation is not just a matter of convenience; it is essential in order to cope with the high throughput of EBS and to help scientists to make informed decisions during the experiment, therefore increasing the chances of collecting scientifically relevant results.
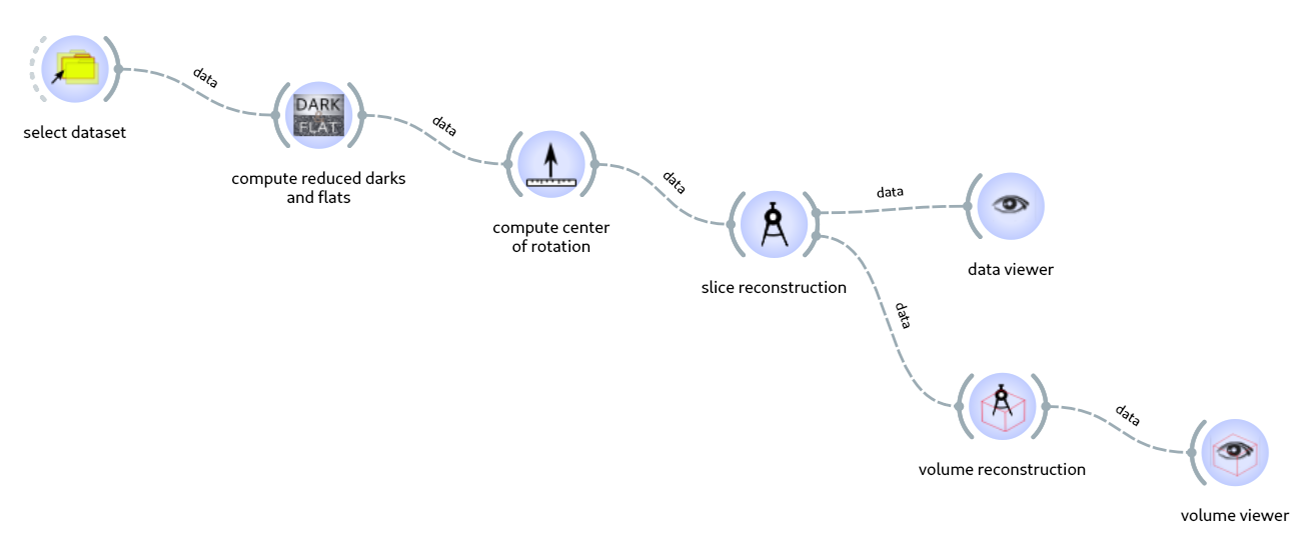
Workflows are data-processing pipelines made of a succession of steps executing specific tasks. Figure 1 shows an example of a workflow. These tasks, which are written by data-analysis experts drawing on their knowledge and experience, can be reused and rearranged by non-expert users to create data-processing pipelines tailored to the experiment and/or beamline. Therefore, workflows combine robustness and flexibility, making them ideal candidates for running automated data processing.
Click image to enlarge
Fig. 1: Example of a tomography workflow.
To support workflow execution and the data-processing automation that comes with it, the ESRF Workflow System, or EWOKS, was developed by the Data Automation Unit (DAU) of the Software group. The added value of EWOKS lies in the fact that it is not limited to a single execution workflow system but is instead able to integrate any workflow framework. This means existing workflows, such as those used in structural biology or tomography, are easily integrated into EWOKS. Moreover, this makes EWOKS much more resilient and flexible since it is, by design, isolated from the underlying software technologies used to execute and manage workflows.
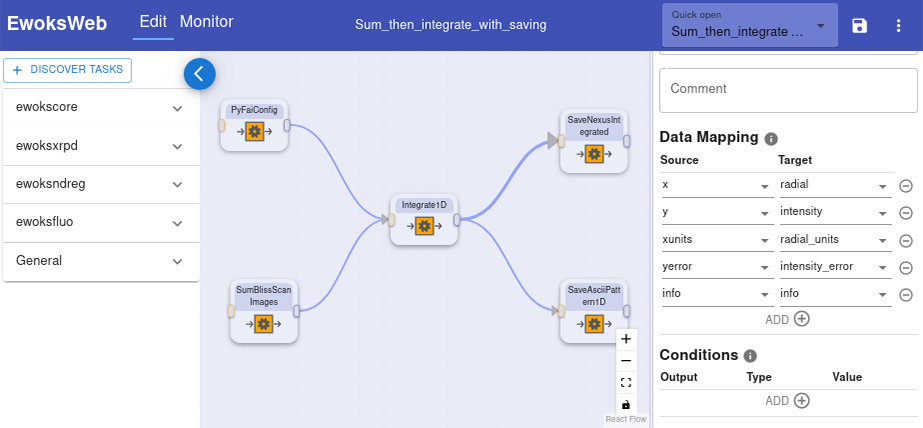
EWOKS can be installed locally by users and can also be deployed as a centralised service on beamlines. This allows EWOKS to be used either for local processing or for automated processing on the beamline. For further ease of use, EWOKS also comes with a web editor to create and edit workflows (Figure 2).
Click image to enlarge
Fig. 2: EWOKS web editor with a loaded workflow.
To better integrate with existing scientific tools, EWOKS is written in Python, the language that is now a standard for scientific computing. EWOKS workflows can therefore use any Python code as tasks, including code calling external programs (PyTorch, Fortran, etc.) and Jupyter notebooks, a popular tool for scientific data processing. This makes EWOKS a growing ecosystem where scientists can pick from more than 400 tasks to build data-processing pipelines [1]. This ecosystem is catered and extended by the DAU team, that works closely with the beamlines to deliver the most efficient data-processing experience possible.
Thanks to its high flexibility, EWOKS can interact with BLISS, the ESRF beamline data acquisition system. This makes it possible to trigger workflows as soon as the data are acquired, enabling a wide range of possibilities as the acquired data can be processed and reduced automatically, without any input from the user. EWOKS workflows are currently in operation on 17 beamlines at the ESRF, with eight more beamlines planned by mid-2024, including tomography, scattering, spectroscopy and macromolecular crystallography beamlines. In the latter case, workflows are not only used for processing but to automatically pilot the beamline, testifying to the robustness of EWOKS.
EWOKS also plays an important role in making data processing reproducible. The triggered workflows can be easily saved and reused by users who want to reprocess data independently at home. Once saved, workflows can also serve as a data-provenance document for scientific publications to describe the data reduction process. EWOKS can also automatically upload processed datasets to the ESRF data portal [2], to make results Findable, Accessible, Interoperable and Reusable (FAIR) by other users [3]. Moreover, users will soon be able to launch EWOKS workflows from the Data Portal to process or reprocess datasets with a single click.
EWOKS is partly funded by STREAMLINE, a Horizon2020-funded project that will complement the ESRF-EBS upgrade by enhancing user experience through new procedures and systems [4].
References
[1] https://ewokspr.readthedocs.io/
[2] https://data.esrf.fr/
[3] https://www.go-fair.org
[4] https://streamline.esrf.fr