Double mesh scans enable optimised multi-crystal experiments at ESRF MX beamlines
Recent developments in multi-crystal data collection have introduced software packages capable of detecting individual crystals, determining their two-dimensional (2D) positions on the sample holder, and estimating their dimensions through X-ray mesh (raster) scans. These advancements have been successfully implemented at ESRF’s macromolecular crystallography (MX) beamlines.
Share
Two-dimensional X-ray mesh (raster) scans are widely used in MX to identify suitable positions on the sample holder for diffraction data collection. Traditionally, these single mesh scans have formed the basis of the ‘Mesh & Collect’ method, which enables the collection of small angular wedges of diffraction data from multiple crystals identified within the scanned area.
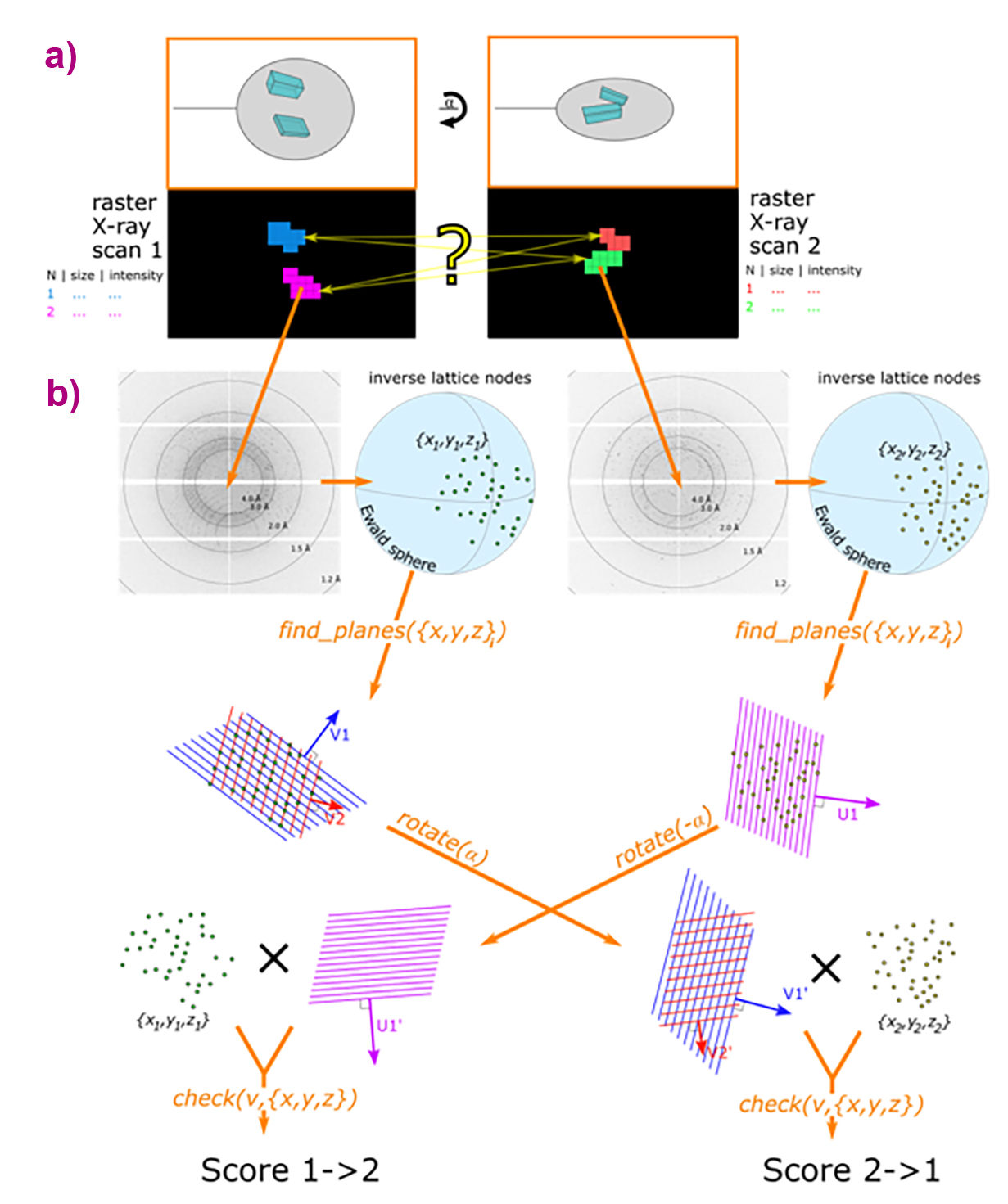
This approach has now been extended by introducing a pair of 2D mesh scans (double mesh scans), performed at different orientations and analysed automatically using dedicated software (Figure 1). The new suite of tools, including Dozor, Dozor-m2 and Resheteau, builds upon the established procedure for determining the 2D positions of individual crystals in individual mesh scans. The key innovation lies in the ability to identify correspondences between crystal areas in the two scans, enabling optimal three-dimensional (3D) centring and beam size selection for multi-crystal data collection.
Click figure to enlarge
Fig. 1: Workflow of the ‘Double Mesh & Collect’ method. (a) Two mesh scans are performed over the sample area at different orientations of the rotation axis, separated by an angular difference α. Analysis with Dozor and Dozor-m2 identifies two crystals (shown in different colours) within each mesh scan. The yellow question mark highlights ambiguity in selecting the optimal crystal centring coordinates. (b) Schematic of the Resheteau algorithm. Raw diffraction images are first processed by Dozor to extract spot coordinates. These coordinates are then used by Resheteau to reconstruct lattice subsets in reciprocal space. Finally, Resheteau compares scores from different plane vectors, with the maximum score providing the final match between crystals in the two scans.
A major enhancement is delivered by the Resheteau algorithm, which recognises identical crystals across different mesh scans by analysing their diffraction patterns at known orientations. Previously, this task required indexing algorithms, which are inefficient when applied to two still images from separate scans. Resheteau demonstrates high efficiency at medium resolutions (2.0–3.0 Å), though its performance decreases for poorly diffracting crystals with resolution worse than 3.0 Å, where fewer diffraction points reduce the reliability of analysis.
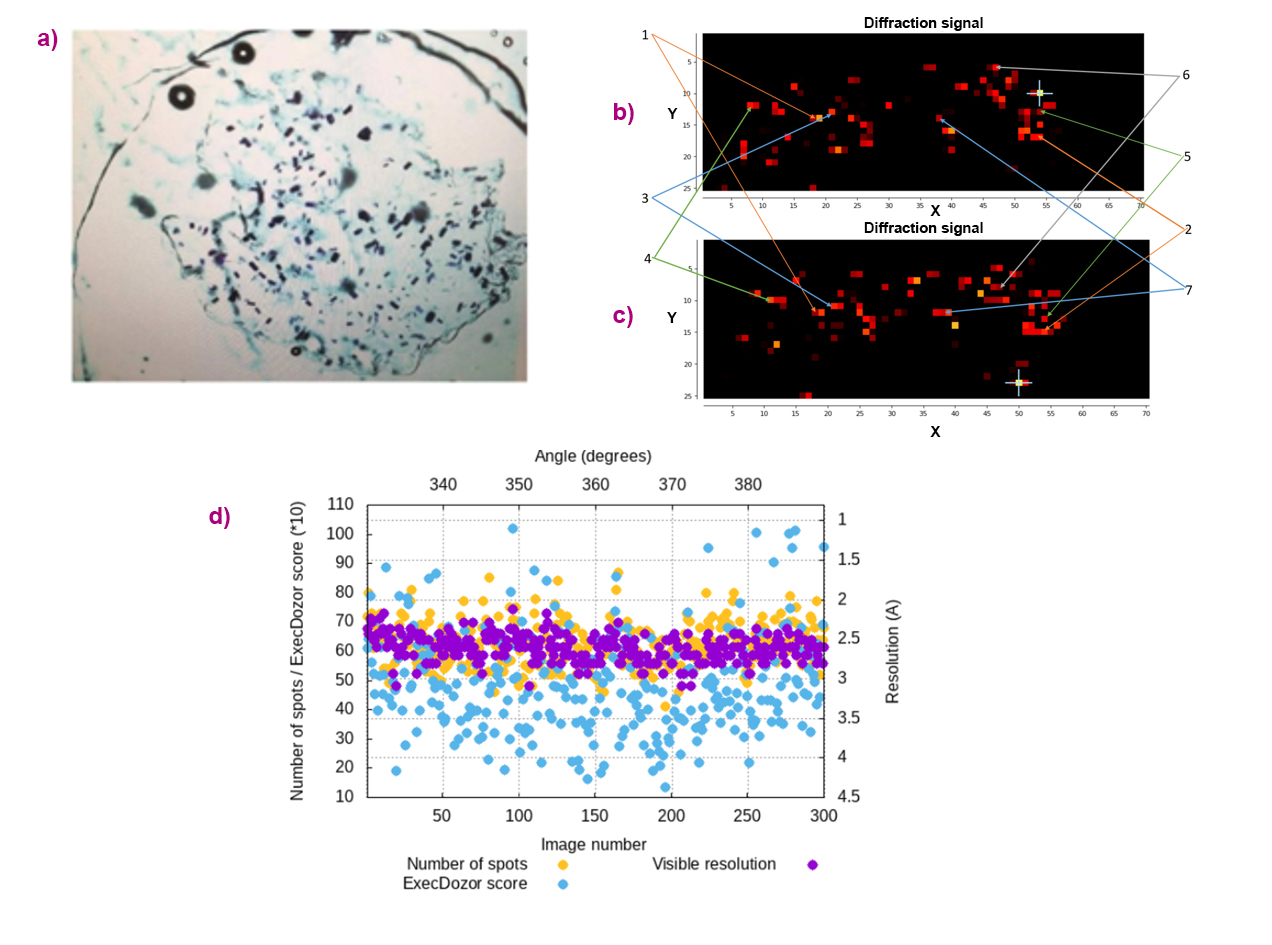
The new ‘Double Mesh & Collect’ method offers several practical advantages. It enables the collection of diffraction wedges as wide as 360° per crystal. In addition, the required sample rotation between the two mesh scans can be substantially smaller than 90°, which is particularly valuable in MX experiments with limited rotation ranges, such as measurements using crystallisation plates or chips (Figure 2). By providing accurate 3D positional information for all crystals on the sample holder, this approach facilitates the rational organisation of fully automated multi-crystal data collection.
Click figure to enlarge
Fig. 2: Multi-crystal data collection from crystals of the membrane protein BR. (a) Snapshot of the crystalline sample sandwiched between two Mylar foils. (b,c) Diffraction heat maps from two mesh scans, separated by 60° of sample rotation. Crystal maps generated by Dozor-m2 for each mesh scan. Crystals from which data have been collected are indicated by arrows. (d) Data collection plots for one selected crystal, showing the angular coverage of the collected diffraction wedges.
This methodology represents a significant advancement in MX sample characterisation. By maximising data quality and quantity from each sample, ‘Double Mesh & Collect’ enhances the efficiency and automation of multi-crystal experiments, thereby supporting more comprehensive and reliable structural studies.
Principal publications
Enhanced capabilities for multi-crystal data collection based on double mesh scans, I. Melnikov et al., Acta Crystallogr. D Struct, Biol. 81(7), 344-356 (2025); https://doi.org/10.1107/S2059798325005145
The complex analysis of X-ray mesh scans for macromolecular crystallography, I. Melnikov et al., Acta Crystallogr. D Struct. Biol. 74(4), 355-365 (2018); https://doi.org/10.1107/S2059798318002735