- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2016
- Structural biology
- Structural insights into tRNA wobble base modification reaction by the Elongator complex
Structural insights into tRNA wobble base modification reaction by the Elongator complex
The Elongator complex is involved in eukaryotic cells protein manufacture. The crystal structure of Elongator complex subunit Elp3 was determined here, providing new insights into the arrangement and interplay of the two Elp3 domains within Elongator and stimulating additional biochemical, biophysical and phenotypical analyses. These allowed the tRNA binding site to be mapped and a regulatory loop was identified that seems to link tRNA binding and acetyl-CoA hydrolysis.
The synthesis of proteins is tightly controlled to assure their production in the right place and at the right time. Incorrectly-folded proteins, inappropriate expression levels or the production of a protein in the wrong cellular context leads to cellular dysfunction and can cause severe diseases and cancer in the worst case. Cells employ a multi-subunit complex, called Elongator, to attach chemical modifications to tRNAs. This guarantees that proteins are produced with the highest precision and at the correct speed.
The highly-conserved eukaryotic Elongator complex has been associated with a plethora of cellular activities, but it is now widely accepted that its main cellular function is the formation of 5-methoxycarbonylmethyl-uridine (mcm5U) in the wobble base position of eukaryotic tRNAs [1]. Such uridine modifications have been shown to enhance tRNA binding to the aminoacyl site of the ribosome, affect the speed of translating ribosomes and secure the homeostasis of whole cellular proteomes. Among its six individual subunits, the Elongator complex Elp3 has been predicted to contain a unique combination of a KAT domain and a radical SAM binding domain. As these two well-known domains could provide the suitable radical intermediates necessary to perform the respective modification reaction, they have been suspected to constitute the active site of Elp3. We have recently determined crystal structures of other Elongator subunits [2] and regulatory factors [3], but, as no high-resolution structural information for the enzymatically active subunit was available, the details of the uridine modification reaction remained elusive.
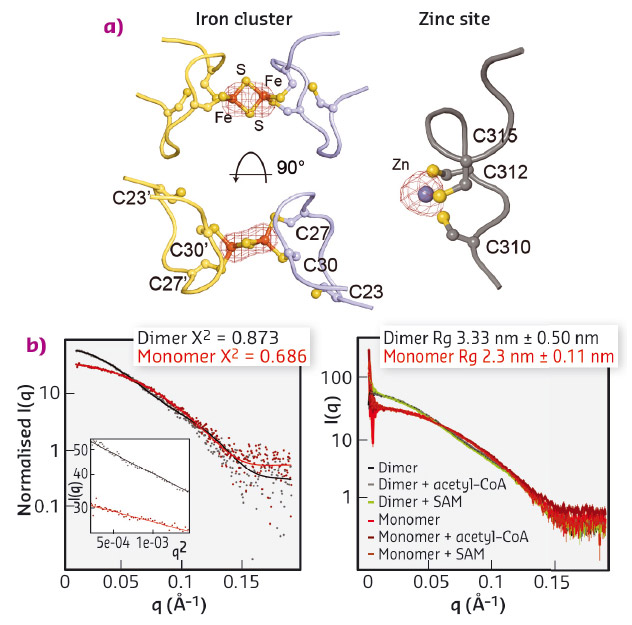
We expressed and purified large quantities of Elp3 from Dehalococcoides mccartyi (DmcElp3), an extremophilic anaerobic bacterium recently isolated from toxic waste. Subsequently, we managed to crystallise the protein and collect diffraction data at beamline ID29 to an overall resolution of 2.15 Å, allowing us to solve the structure using the single-wavelength anomalous dispersion (SAD) phasing technique. As the purified protein and the resulting crystals showed a characteristic reddish colour, we suspected that the protein carries an iron sulfur cluster, typical of radical SAM domain proteins. We thus used X-ray fluorescence (XRF) analyses, also available at ID29, to confirm the presence of iron atoms in our crystals and located the cluster by collecting a complete data set close to the absorption edge of iron (λ = 1.7377 Å). Anomalous difference Fourier maps clearly indicated the type of cluster, namely [2Fe2S], as we specifically identified peaks for two coordinated iron ions. In addition, we used XRF analysis and anomalous difference Fourier maps collected at the zinc absorption edge (λ = 1.28238 Å) to locate and characterise a novel and unexpected zinc coordination motif in DmcElp3 (Figure 82a). Last but not least, we observed that the iron sulfur cluster is localised in the interface between two DmcElp3 monomers, something never observed before in any known radical SAM domain. We then used small-angle X-ray scattering (SAXS) at beamline BM29 to confirm that the dimer exists in solution and that it is unchanged in the presence of specific ligands (Figure 82b).
 |
|
Fig. 82: a) Identification of iron (left) and zinc (right) sites using anomalous difference Fourier maps calculated from datasets recorded close to the respective absorption edges. b) Fitting of experimental SAXS data (dots) for DmcElp3 (monomer/dimer) to theoretical scattering curves calculated from the crystal structure (left). SAXS analyses of DmcElp3 in the presence and absence of acetyl-CoA and SAM (right). |
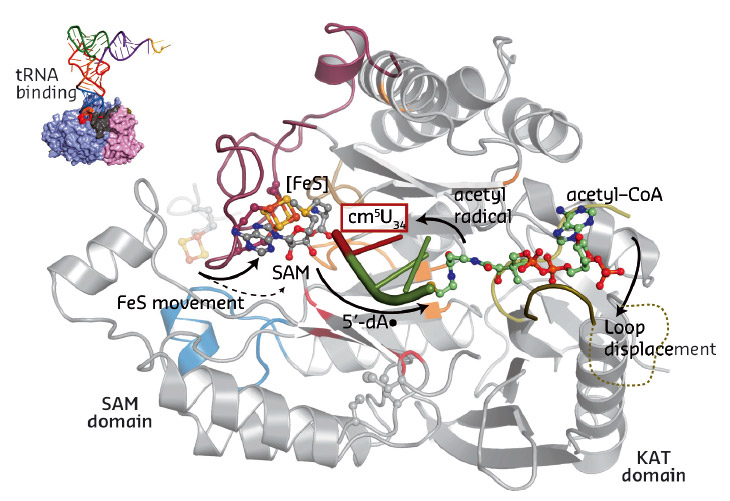
Our work for the first time provides a structural framework for the previously proposed radical based modification reaction (Figure 83). Strikingly, the obtained structural data allowed us to locate most of the previously-published disease-related mutations. Subsequently, we tested the identified structure-guided and disease-related mutations in vivo and showed that they indeed affect the tRNA modification activity of Elongator, thus suggesting a direct correlation between Elp3 dysfunction and disease progression.
 |
|
Fig. 83: Summary of the individual steps of the proposed radical based tRNA modification reaction. The two Elp3 domains, the iron sulfur cluster, SAM, acetyl-CoA and the bound tRNA anti-codon loop are highlighted. |
Principal publication and authors
Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi, S. Glatt (a,b), R. Zabel (c), O. Kolaj-Robin (d,e,f), O.F. Onuma (c), F. Baudin (a,g), A. Graziadei (a), V. Taverniti (d,e,f), T.-Y. Lin (b), F. Baymann (h), B. Séraphin (d,e,f), K.D. Breunig (c) and C.W. Müller (a), Nat Struct Mol Biol. 23, 794–802 (2016); doi: 10.1038/nsmb.3265.
(a) European Molecular Biology Laboratory (EMBL), Heidelberg (Germany)
(b) Max Planck Research Group at the MCB/JUK (Poland)
(c) Martin-Luther-University Halle-Wittenberg (Germany)
(d) University Strasbourg, IGMBC (France)
(e) CNRS, IGMBC (France)
(f) Inserm, IGMBC (France)
(g) University Grenoble Alpes–CNRS–EMBL (France)
(h) Laboratoire de Bioénergétique et Ingénierie des Protéines (France)
References
[1] S. Glatt and C.W. Müller, Curr Opin Struct Biol. 23, 235-42 (2013).
[2] S. Glatt et al., Nat Struct Mol Biol. 19, 314-20 (2012).
[3] S. Glatt et al., Structure 23, 149-60 (2015).



