- Home
- News
- Spotlight on Science
- An original Cu(I)-coordination...
An original Cu(I)-coordination shell in a small bacterial metalloprotein identified by XAS and NMR spectroscopy
25-06-2010
The combined use of XAS and NMR spectroscopy enabled characterisation of the three-dimensional structure and the metal-binding site of a small Cu(I)/Cu(II)-binding protein involved in bacterial heavy-metal resistance. This protein features a binding cooperativity with enhanced affinity for Cu(II) once Cu(I) is bound. NMR-derived data were used for the determination of the atomic coordinates of the protein moiety in solution while XAS measurements yielded geometric information on the Cu(I) site which is coordinated by four sulphur atoms. This study represents the first example of a XANES and EXAFS characterisation of a tetrathioether Cu(I) site in a protein in which copper is not part of a cluster. Knowledge of this new structure permits the understanding of the previously observed Cu(I)/Cu(II) binding cooperativity from a structural point of view.
Share
Living organisms are continuously exposed to environmental stress originating from sources such as toxic compounds, radiation and high temperature. During evolution, they have adapted to the specific environment in which they live and have developed resistance mechanisms to protect themselves against stress events. One well-known class of toxic products is formed by heavy metals that naturally occur in the environment but are also released in huge quantities through anthropogenic processes, especially since the beginning of industrialisation. Cupriavidus metallidurans CH34 is a bacterium that was isolated from a zinc decantation tank in Belgium. It can resist high concentrations of many heavy metals (Cu(II), Zn(II), Cd(II), Co(II), Pb(II), Hg(II), Ni(II), Ag(I) and Cr(VI)). This strain became a model for studying metal resistance mechanisms in microorganisms. In our study we were interested in the molecular and metal binding properties of proteins involved in heavy metal resistance in this bacterium.
Many vital processes require copper as a redox-active cofactor but excess copper damages cells by catalysing the production of free radicals. Therefore, several mechanisms have evolved which maintain a suitable level of intracellular copper and control its oxidation state. Copper resistance of C. metallidurans CH34 involves more than 19 proteins that are produced in response to excess copper. One of these is CopK, a small periplasmic protein that is produced in abundance under copper stress. As a small metal binding protein probably involved in metal trafficking, it is called a metallochaperone. We have previously determined the solution structure of apo-CopK [1] and it has also been shown that CopK binds Cu(I) and Cu(II) in a cooperative way [2].
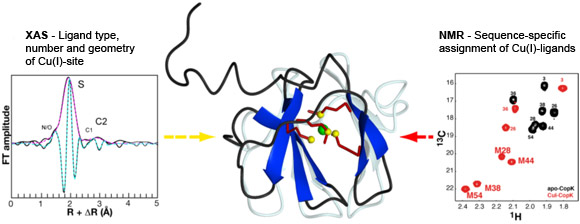
A collaboration of researchers from the NMR and the Membrane Protein groups at the Institut de Biologie Structurale, the Environmental Geochemistry Group at the Université Joseph Fourier, the Institut Néel and beamline BM30B (FAME CRG) succeeded in the determination of the solution structure of the Cu(I)-bound CopK protein and the characterisation of its Cu(I)-site. A so far unique Cu(I) ligand sphere was discovered that is formed by four thioether groups belonging to four methionines from the same protein chain. Spectroscopic data obtained by Nuclear Magnetic Resonance (NMR) and by X-ray Absorption (XAS) were combined for the detailed characterisation of Cu(I)-CopK and its metal site (Figure 1). NMR provides information on intramolecular proton-proton distances that yield the atomic coordinates of the protein chain, the three-dimensional protein structure. However, the Cu(I) ion is NMR-silent and no geometric information on the metal site can be obtained. A cryo-cooled protein solution containing Cu(I)-CopK was therefore studied by XAS, and Cu K-edge XAS spectra were collected at beamline BM30B. The resulting XANES and EXAFS spectra were in accordance with a ligand sphere formed by sulphur atoms and the fit of the EXAFS data suggested the presence of four sulphur atoms in the Cu(I) site. This result was surprising as most small periplasmic bacterial Cu(I)-binding metallochaperones contain Cu(I) bound by sulphur and nitrogen ligands. In addition, a tetrathioether Cu(I) site has so far only been observed in a single protein in which the Cu(I) ion belongs to a copper-molybdenum (MoS2CuS2Mo) cluster. However, a tetrathioether Cu(I) site was also in accordance with a Cu(I)-S distance of 2.31 Å as determined by EXAFS. This distance is very untypical for a trigonal Cu(I) site. These findings were further corroborated by the observation of chemical shift differences in 1H-13C correlation NMR spectra occurring upon Cu(I)-binding. Four peaks, corresponding to methionine methyl groups were significantly affected by the addition of Cu(I) to the protein. This observation leads to the suggestion that the chemical shift of these carbon atoms is a new and interesting probe for the detection of methionine-bound Cu(I) and, most importantly, allows the sequence-specific identification of the Cu(I) ligands.
The structure of Cu(I)-CopK bridges the gap between what was previously known on the Cu(I)/Cu(II)-binding cooperativity and the structures published beforehand. Knowledge of this new structure is the basis for the understanding of the observed Cu(I)/Cu(II) binding cooperativity from a structural point of view. We have demonstrated that the accommodation of Cu(I) in the tetrathioether site is associated with an important structural modification of the C-terminal part of the protein and propose that this reorientation is required for the formation of the Cu(II)-specific site. This explains the much higher Cu(II) affinity of Cu(I)-CopK compared to apo-CopK.
References
[1] B. Bersch, A. Favier, P. Schanda, S. van Aelst, T. Vallaeys, J. Covès, M. Mergeay and R. Wattiez, J. Mol. Biol. 380, 386-403 (2008).
[2] L.X. Chong, M.-R. Ash, M.J. Maher, M.G. Hinds, Z. Xiao and A.G. Wedd, J. Am. Chem. Soc. 131, 3549-3564 (2009).
Principal publication and authors
G. Sarret (a), A. Favier (b), J. Covès (b), J.-L. Hazemann (c), M. Mergeay (d), B. Bersch (b), CopK from Cupriavidus metallidurans CH34 binds Cu(I) in a tetrathioether site: characterization by X-ray absorption and NMR spectroscopy, J. Am. Chem. Soc. 132, 3770-3777 (2010).
(a) LGIT, Grenoble (France)
(b) Institut de Biologie Structurale, Grenoble (France)
(c) Intitut Néel, Grenoble (France)
(d) SCK-CEN, Mol (Belgium)
Top image: XAS and NMR studies of the CopK protein involved in bacterial copper resistance reveal a tetrathioether Cu(I)-site.