- Home
- News
- Spotlight on Science
- Engineering the...
Engineering the substrate binding site of the hyperthermostable archaeal endo‑β‑1,4‑galactanase from Ignisphaera aggregans
28-02-2022
X-ray crystallography data collected at beamline ID23-2 has revealed the structure of the hyperthermostable endo-β-1,4-galactanase from Ignisphaera aggregans. The structure points to the features responsible for hyperthermostability and shows unique features of the substrate binding site among structurally characterised galactanases.
Effective conversion from biomass to biofuel and other bio-derived products can be accomplished by enzymatic degradation followed by fermentation or extraction. One of the many enzymes needed for a full degradation of plant material is the endo-β-1,4-galactanase, which degrades the galactan and arabinogalactan found in pectin, a highly abundant constituent in the primary cell wall of non-woody plants [1].
Thanks to the focused microbeam at the ID23-2 beamline, the structure of the hyperthermostable endo-β-1,4-galactanase from Ignisphaera aggregans (IaGal) was determined from a multiple needle crystal. This is the most thermostable galactanase characterised to date, with a temperature activity optimum of 95ºC and melting temperature of 105ºC, making it highly interesting from an industrial point of view. Combined with previously solved galactanase structures, this study found that cation-π and π-π interactions, including a highly conserved clustered of π-π interactions, are the structural features most correlated to thermostability in this family of enzymes.
The structure of IaGal also revealed a unique length of the substrate binding site compared to other structurally determined galactanases to date. In polysaccharide-degrading glycoside hydrolases such as galactanases, subsites refer to the binding pockets on the enzyme. Each subsite can bind a sugar ring of the polysaccharide, and subsites are often rich in aromatic residues, especially Trp. While fungal galactanases from origins such as Aspergillus aculeatus (AaGal) contain two binding subsites towards the non-reducing end of the substrate [2], and a bacterial galactanase from Bacillus licheniformis (BlGal) contains four [3], IaGal is an intermediate containing three binding subsites (Figure 1) at the non-reducing end.
The number of binding subsites is heavily related to the length of the degradation products produced by the enzyme. Short products are both preferred by the fermenting organisms and lower the viscosity of the resulting biomass slurry and thereby ease liquid handling. However, longer products can be useful when using the degradation products as building blocks for sustainable biomaterials and health products. The researchers therefore attempted to increase the applicability of IaGal by creating rationally designed subsite-deleted variants containing two binding subsites and rationally designed subsite-extended variants containing four binding subsites (Figure 1).
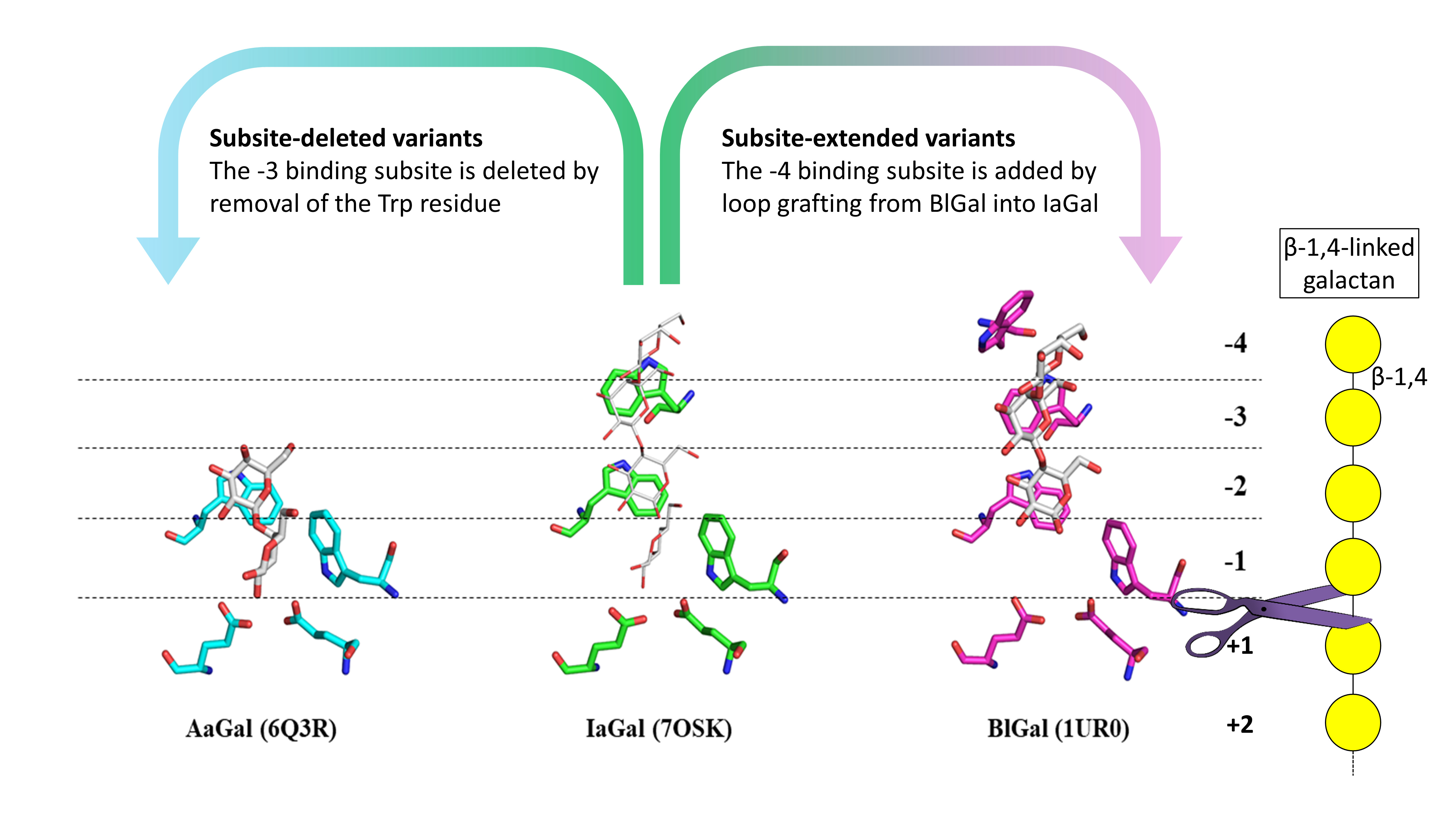
Fig. 1: Substrate binding sites of AaGal, IaGal and BlGal. Substrate subsites at the non-reducing end increase incrementally from fungal enzymes (AaGal PDB ID 6Q3R) to archaeal enzymes (IaGal PDB ID 7OSK) to the bacterial BlGal enzyme (PDB ID 1UR0). The galactobiose and galactotriose ligands are shown as white sticks in the AaGal and BlGal structures, respectively. The same ligands are shown as white lines after superposition onto the IaGal structure. β-1,4 galactan is shown with the galactose monomers indicated by a yellow circle and the glycosidic bonds are indicated by a black line. The enzymes cut the substrate between the -1 and the +1 binding subsite as indicated with a scissor.
The created variants retained very high thermostability. Homology models of these variants were subjected to molecular dynamic simulations to ensure the mutagenesis had not introduced unwanted structural interactions or flexibility. Remarkably, the subsite-extended variants, which contain a rather sizeable additional loop, show no increase in fluctuation of the added binding subsite.
The degradation profiles of the variants, the wild type IaGal and reference galactanases were obtained by enzymatic degradation of lupin galactan and measuring the products using high-performance anion exchange chromatography coupled with pulsed amperometric detection. This technique enables precise quantification of the oligosaccharide degradation products. These results show that the degradation pattern of the subsite-deleted variants successfully resembles the degradation pattern of the reference galactanase Aagal containing only two binding subsites. The subsite-extended variant obtains a unique degradation pattern compared to both the reference galactanases and the wild type IaGal (Figure 2).
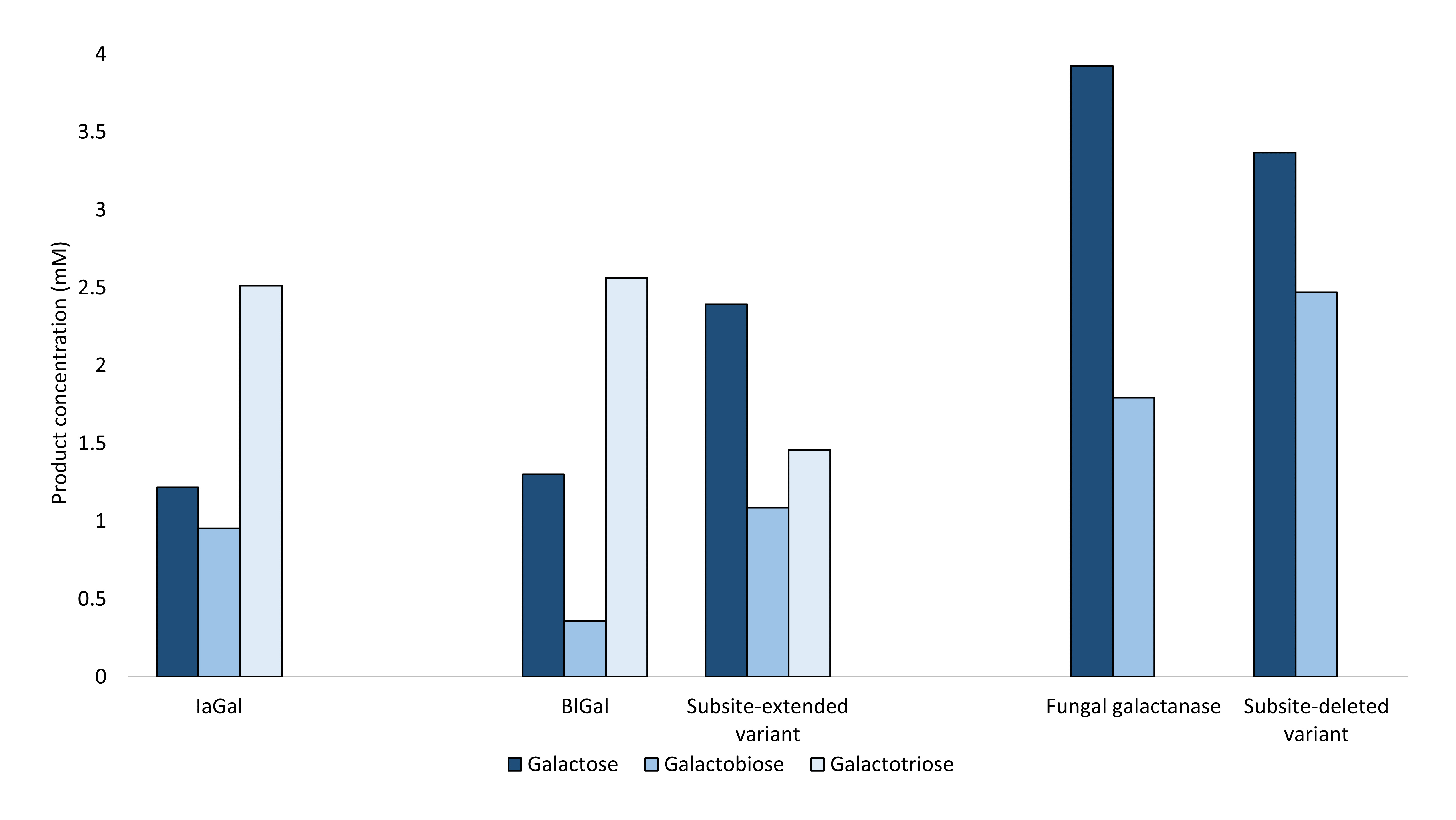
Fig. 2: The substrate degradation pattern at full hydrolysis for IaGal, a subsite-deleted variant, a subsite-extended variant and reference WT galactanases – the fungal, and the bacterial BlGal. The variants shown are representative for other variants tested. The bars represent the degradations products measured in mM.
Taken together, this research indicates that engineering of the substrate binding site of hyperthermostable enzymes towards certain products can be a feasible protein engineering strategy as an alternative to introducing thermostability features in enzymes with the desired product patterns.
Principal publication and authors
Engineering the substrate binding site of the hyperthermostable archaeal endo‑β‑1,4‑galactanase from Ignisphaera aggregans, S.J. Muderspach (a), F. Fredslund (a,d), V. Volf (b,e), J.C.N. Poulsen (a), T.H. Blicher (b), M.H. Clausen (c), K.K. Rasmussen (a), K.B.R.M. Krogh (b), K. Jensen (b), L.L. Leggio (a), Biotechnol. Biofuels 14, 183 (2021); https://doi.org/10.1186/s13068-021-02025-6
(a) Department of Chemistry, University of Copenhagen, Copenhagen (Denmark)
(b) Novozymes A/S, Kongens Lyngby (Denmark)
(c) Department of Chemistry, Technical University of Denmark, Kongens Lyngby (Denmark)
(d) The Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, Kongens Lyngby (Denmark)
(e) Department of Genetics, Harvard Medical School, Boston (USA)
References
[1] J.M. Lau et al., Carbohydr. Res. 137, 111-125 (1985).
[2] S. Torpenholt et al., Acta Crystallogr. Sect. F. 75, 399-404 (2019).
[3] C. Ryttersgaard et al., J. Mol. Biol. 341, 107-117 (2004).



