- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2016
- Complex systems and biomedical sciences
- Rotation after red light absorption – the structure of a photoactivated phytochrome revealed
Rotation after red light absorption – the structure of a photoactivated phytochrome revealed
Time-resolved X-ray scattering data from red light sensitive phytochromes provide new insights into the structural changes of the protein complex upon photo activation. These findings have improved our understanding of complex sensory proteins, the manipulation of which may have future applications in agriculture and optogenetics.
Phytochromes are allosterically regulated, photoswitchable two-component signalling proteins that function in plants, fungi and bacteria. They change their conformation, and thereby their biological activity, upon the absorption of red light. In the bacterial species Deinococcus radiodurans, a red/far-red light absorbing phytochrome controls the production of light-protective pigments. In work carried out at beamline ID09 at the ESRF, and at the SLS and APS, structural data was collected on the activation process of the full phytochrome system.
The phytochrome protein complex contains a photo sensory module and a biological effector domain, typically a histidine kinase in bacteria. The experiments used time-resolved X-ray scattering measurements of the phytochrome constructs as they switch between the two states. To achieve this, the phytochrome solution was flashed with short bursts of red light to convert the molecules between the resting and the active state. Scattering data was collected between the flashes. The measurements were performed on both the truncated and the full-length phytochrome system. The truncated versions of the protein contained the so-called chromophore binding domain (PAS-GAF) or the photosensory unit (PAS-GAF-PHY).
 |
|
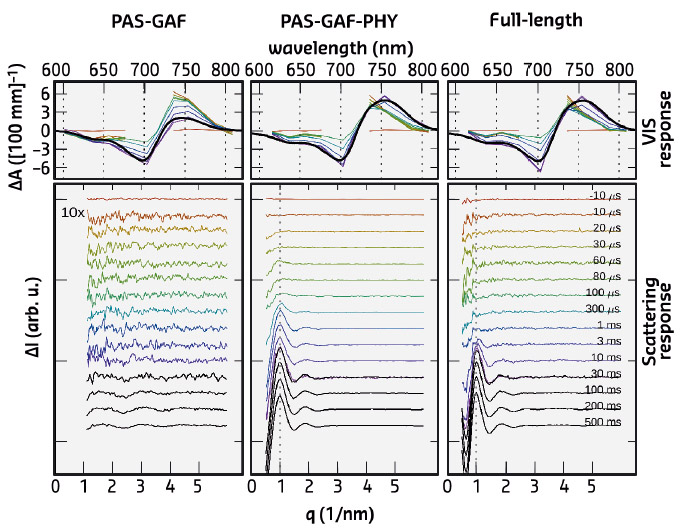
Fig. 102: Time-resolved optical absorption spectra (top) and time-resolved X-ray scattering spectra (bottom). The left panel (PAS-GAF) shows the chromophore binding domain, the middle-panel (PAS-GAF-PHY) shows the data from the photosensory unit, and the right panel (full-length) is from the full phytochrome protein complex. Colours indicate the detection time after the red-light (671 nm) laser flash. |
The X-ray scattering data revealed major signals in the low-q range of the spectra. This reflects the large-scale structural changes in the full-length protein complex and in the photosensory unit. In contrast, the chromophore binding domain, the most compact subunit, revealed minimal structural changes in the scattering. The large-scale changes occur on the ms time scale, while the spectroscopic experiments show characteristic signals in the Vis-absorption spectra on the ns-, ms, and ms-timescale (Figure 102). No fast structural changes were observed by X-ray scattering. By linking the X-ray time constants with the spectroscopic ones, it was shown that the large-scale structural changes are the end-product of the photocycle. Hence, all the early-time states contain small-amplitude structural changes and possibly chemical changes in the protein backbone, but these are too small to be detected.
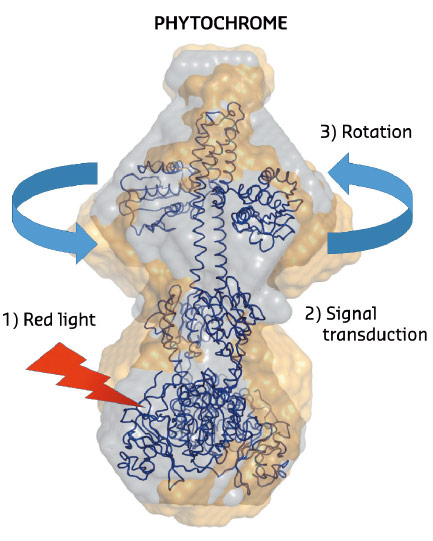
Recently, it was shown that a portion of the truncated photosensory core module (PAS-GAF-PHY) undergoes refolding when it is exposed to red light and that the refolding is associated with a large-scale opening motion [1]. In the present study, ab initio structural modelling of the X-ray data revealed that the histidine kinase effector domain restricts the large-scale movements in the full-length protein. In this case the conformational push from the refolding action is used to generate a screw motion which rotates the effector domain (Figure 103).
 |
|
Fig. 103: The structural changes in phytochrome proteins. The phytochromes absorb red light in the biliverdin molecules located on the lower side of the protein (red arrow). After red light absorption, chemical and structural changes take place in the protein interior (signal transduction). This leads to a rotational movement on the top of the protein which generates signals to other parts of the cell. Grey spheres describe the structure of the protein in the dark state and the yellow spheres describe the structure of the illuminated state. |
To summarise, the scattering data from the photoactive protein phytochrome revealed a rotational movement of the effector domain of the protein. These large-scale structural changes are associated with the last steps of the photocycle of the phytochrome. How these large scale movements are linked to the chemical reactions in the output domain is to be shown in future studies.
Principal publication and authors
Structural photoactivation of a full-length bacterial phytochrome, A. Björling (a), O. Berntsson (a), H. Lehtivuori (b), H. Takala (b), A.J. Hughes (a), M. Panman (a), M. Hoernke (a), S. Niebling (a), L. Henry (a), R. Henning (c), I. Kosheleva (c), V. Chukharev (d), N.V. Tkachenko (d), A. Menzel (e), G. Newby (f), D. Khakhulin (f), M. Wulff (f), J.A. Ihalainen (b) and S. Westenhoff (a), Science Advances 2, e1600920 (2016); doi: 10.1126/sciadv.1600920.
(a) University of Gothenburg (Sweden)
(b) University of Jyväskylä (Finland)
(c) The University of Chicago (USA)
(d) Technical University of Tampere (Finland)
(e) Paul Scherrer Institut (Switzerland)
(f) ESRF
References
[1] H. Takala et al., Nature 509, 245 (2014).



