- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2015
- Structural biology
- Cellular sodium and potassium pumps light up optogenetics
Cellular sodium and potassium pumps light up optogenetics
The structure of a light-activated ion pump has been solved. This discovery provides new tools in the emerging field of optogenetics, which has potential long-term applications such as vision and hearing regeneration.
Microbial rhodopsins are a large family of photosensitive membrane proteins that use light to transport ions into and out of the cell, generating phototaxis signals that cause the cell to move towards favourable illumination conditions. The most studied system, bacteriorhodopsin, was discovered in 1971 but thousands more have since been identified in microorganisms as diverse as bacteria, algae and viruses, exhibiting various light-gated channels and light-driven proton and anion pumps. Incorporating these proteins into neurons has allowed precise control of neural impulses and lies at the foundation of the growing field of optogenetics. So far, however, all the known light-driven pumps are unable to transport sodium and potassium – the ions whose motions across the cell membrane constitute natural neural impulses.
While investigating the marine bacterium Krokinobacter eikastus, K. Inoue and co-workers [1] unexpectedly discovered that K. eikastus cells are able to pump out sodium ions during early periods of growth. It turned out that the corresponding ion transporter, dubbed KR2, belongs to a new type of microbial rhodopsin in which some of the most important ionisable amino acids are replaced with polar ones, and vice versa. Even when expressed in another commonly employed bacterium, E. coli, KR2 could still transport sodium ions under illumination. Yet it appeared that potassium ions, despite being very similar to sodium ions, were not transported by KR2.
 |
|
Fig. 120: The crystals of KR2 used for structure determination. |
This and other curiosities displayed by KR2 attracted our interest because we had already carried out several structural studies of other microbial rhodopsins to investigate the molecular mechanisms behind their actions. However, membrane proteins are notoriously difficult to handle and producing them is usually expensive and time-consuming, and KR2 is no exception and the protein could be produced only in batches of several milligrams. Thus, while thousands of crystallisation conditions should have been tested, we had to miniaturise and automate this process using the robotic and imaging systems available at the IBS. The diffraction signal from the first crystals obtained, which measured less than 20 μm across, was too weak to be measured using in-house X-ray sources. Recent advances at the ESRF’s macromolecular crystallography beamlines – such as highly brilliant microfocus X-ray beams, automatic sample changers and convenient software protocols – greatly helped the search for the best crystals. After several months of trials at beamlines ID23-1 and ID29, we were able to improve crystal size (Figure 120) and a resolution of the diffraction data collected from around 20 Å to better than 1.5 Å. Meanwhile we were also able to extract information about KR2’s spectroscopic characteristics using optical absorption spectra collected at the cryobench laboratory (beamline ID29S).
 |
|
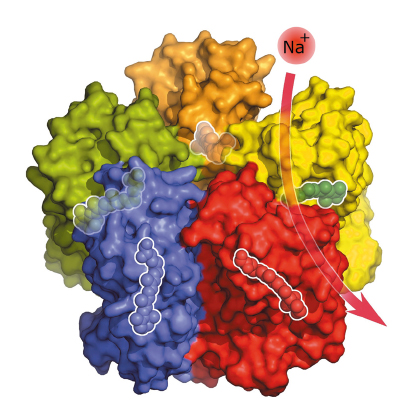
Fig. 121: Under physiological conditions, five KR2 molecules spontaneously form a star-shaped complex with each KR2 molecule being able to bind and transport a sodium ion across the membrane. Inside the proteins are highlighted the light-sensitive retinal cofactors that enable the pumping activity. |
The crystal structure of the KR2 sodium pump (Figure 121) revealed a short protein helix that caps the outfacing opening of the pump like a lid. Another interesting feature of KR2 is the unusual structure of the inward facing ion uptake cavity, which is unexpectedly large and protrudes from the protein surface. Hypothesising that this feature could act as a filter that allows KR2 to be selective for sodium ions, we swapped specific amino acids at the relevant site using targeted mutations and found that KR2 indeed loses its sodium-pumping ability under such conditions. But we also found that one mutation transforms KR2 into a light-driven potassium pump – the first of its kind.
For potential optogenetic applications, this result is especially interesting because transporting potassium ions out of the cell is a natural neuron deactivation mechanism. Normally, activated neurons release these ions through passive potassium channels in the membrane, but a light-activated potassium pump would allow this process to be precisely controlled and provide a highly effective off-switch for neurons [2, 3]. The next step is to find ways of integrating the pump into different types of cells. In combination with the light-activated Channelrhodopsin 2, which is a well-known molecular on-switch, the KR2 potassium pump would form a perfect pair of tools for the precise control of nerve cell activity.
Principal publication and authors
Crystal structure of a light-driven sodium pump, I. Gushchin (a,b,c), V. Shevchenko (b,c,d), V. Polovinkin (a,b,c), K. Kovalev (b,c), A. Alekseev (b,c), E. Round (c), V. Borshchevskiy (b,c), T. Balandin (c), A. Popov (e), T. Gensch (c), C. Fahlke (c), C. Bamann (f), D. Willbold (c,g), G. Büldt (b,c), E. Bamberg (f) and V. Gordeliy (a,b,c), Nature Structural and Molecular Biology 22, 390–395 (2015); doi: 10.1038/nsmb.3002.
(a) Institut de Biologie Structurale, Grenoble (France)
(b) Laboratory for Advanced Studies of Membrane Proteins, Moscow Institute of Physics and Technology, Dolgoprudniy (Russia)
(c) Institute of Complex Systems (ICS), Research Center Jülich (Germany)
(d) Institute of Crystallography, University of Aachen (RWTH) (Germany)
(e) ESRF
(f) Max Planck Institute of Biophysics, Frankfurt am Main (Germany)
(g) Institut für Physikalische Biologie, Heinrich-Heine-Universität Düsseldorf (Germany)
References
[1] K. Inoue et al., Nature Communications 4, 1678 (2013).
[2] H. Kato et al., Nature 521, 48–53 (2015); doi: 10.1038/nature14322.
[3] I. Gushchin et al., FEBS Journal (2015); doi: 10.1111/febs.13585.



