- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2016
- X-ray nanoprobe
- X-rays enable a direct view inside biological cells
X-rays enable a direct view inside biological cells
Structures within whole biological cells have been resolved quantitatively at the nanoscale using a combination of ptychography and scanning X-ray nanodiffraction. Employed sequentially, these techniques provided high resolution real space and reciprocal space images while exposing the cells to a comparatively small radiation dose. The structural arrangement and diameter of keratin protein bundles inside epithelial cells was revealed and filament diameters and distances could be derived.
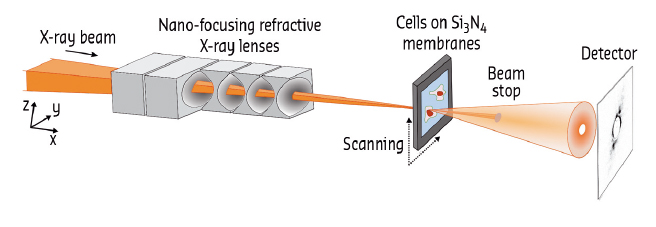
Biological cells contain a plethora of nanometre-sized objects and structures. Much of our knowledge about cellular function has been gained from direct imaging. Traditionally, fluorescence-based techniques and electron microscopy have been used to achieve micrometre to nanometre resolution in cellular imaging. More recently, X-ray imaging methods, which take advantage of the short wavelength and high penetration power of X-rays, were developed and applied to the imaging of biological matter as well [1-3]. The small beams available at specialised synchrotron beamlines such as ID13 (Figure 31) enable high-resolution applications. One of the greatest challenges, however, remains radiation damage by the X-rays, which becomes more pronounced as the resolution reaches smaller values.
 |
|
Fig. 31: Experimental setup at ID13. |
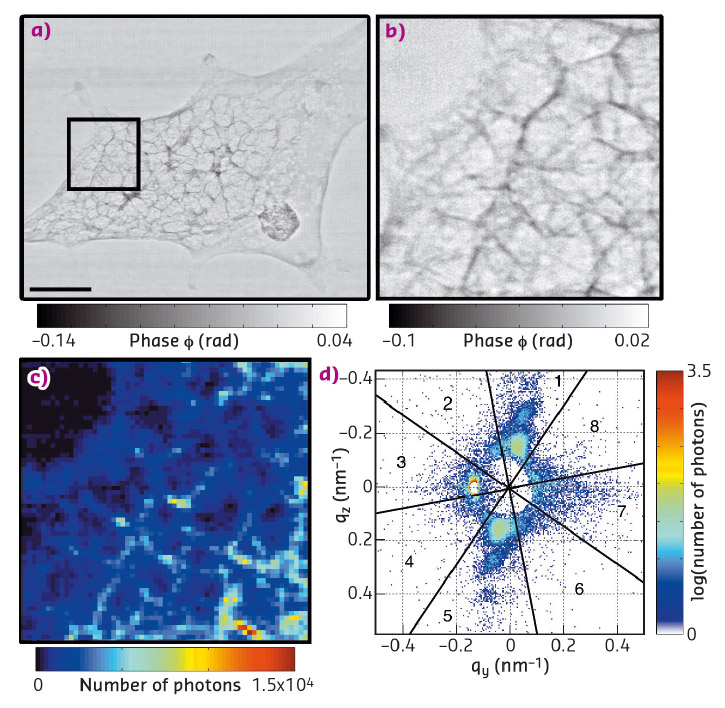
By first recording ptychography overview images of whole cells at 65 nm resolution (Figure 32a and b), cellular structures were located precisely at a comparatively low dose of 103 Gy. Ptychography is a coherent diffractive imaging technique whereby the electron density of the scattering material is determined from a series of two-dimensional scattering patterns via iterative reconstruction algorithms and redundant information from overlapping scanning positions. Subsequently, scanning X-ray nanodiffraction (approximate dose 108 Gy) was employed to record reciprocal space scattering patterns from regions of interest (ROIs). For this technique, the sample was scanned through the beam at a step size comparable to the beam size. The intensity values in the individual scattering patterns were integrated to derive dark-field-images (Figure 32c) at an intermediate resolution, which corresponds to the beam size in the order of 100 nm. Further analysis of each individual pattern revealed a local scattering signal (Figure 32d) which could be assigned to the specific position in the cell.
 |
|
Fig. 32: a) Ptychography image, scale bar 5 μm and b) detail from the ROI shown by the black box in (a). c) Dark-field image computed from scanning X-ray nanodiffraction of the same ROI, and d) example of one pixel from (c) showing an anisotropic signal. |
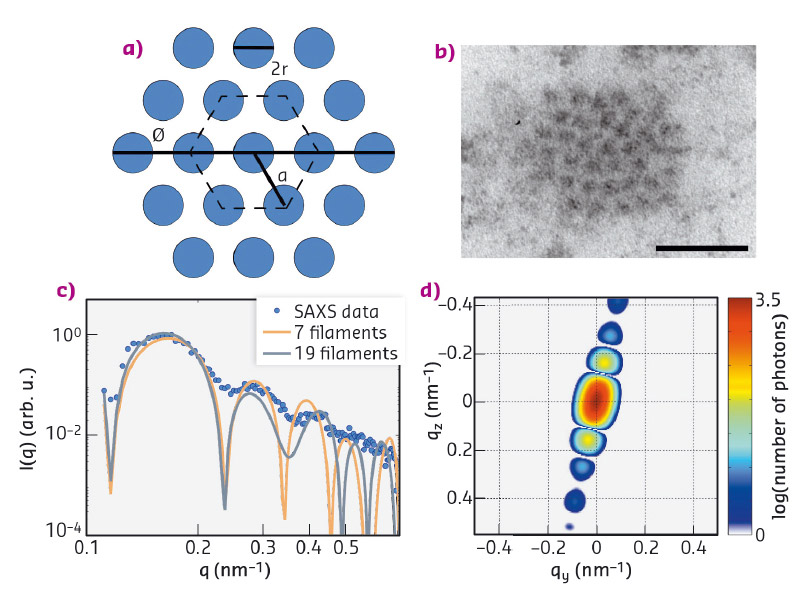
The mechanical properties of biological cells are to a large part governed by the so-called cytoskeleton, a composite network of fibrous proteins [4]. The present study focuses on keratin bundles, which in epithelial cells constitute one of the filament types. By fitting a hexagonally packed bundle model [5] to the data, the radius of individual filaments (r = 5.5 ± 0.7 nm), the inter-filament distance (a = 15.2 ± 1.4 nm) and the bundle diameter (D = 72.0 ± 5.5 nm) could be derived (Figure 33). These values are in agreement with results from electron microscopy [6], but were obtained without slicing or staining the cell and are thus at a reduced risk of artifacts.
 |
|
Fig. 33: a) Cross section of a model of the keratin bundles with 19 filaments per bundle. b) TEM image of the cross section of a keratin bundle in a hexagonal lattice, scale bar 50 nm. c) Radial intensity of the pattern shown in Figure 32d and results from the fit. d) 2D simulated pattern corresponding to the fit with 19 filaments in panel (c). |
This study demonstrates the advantage of combining ptychography and scanning X-ray nanodiffraction in biological imaging since it provides nanometre resolution imaging of specific cellular structures while avoiding pronounced radiation damage of the samples.
Principal publication and authors
X-rays reveal the internal structure of keratin bundles in whole cells, C.Y.J. Hémonnot (a), J. Reinhardt (b), O. Saldanha (a), J. Patommel (c), R. Graceffa (a), B. Weinhausen (d), M. Burghammer (d,e), C.G. Schroer (b,f) and S. Köster (a), ACS Nano 10, 3553-3561 (2016); doi: 10.1021/acsnano.5b07871.
(a) Institute for X-ray Physics, University of Göttingen (Germany)
(b) DESY, Hamburg (Germany)
(c) Institute of Structural Physics, TU Dresden (Germany)
(d) ESRF
(e) Department of Analytical Chemistry, Ghent University (Belgium)
(f) Institute for Nanostructure and Solid State Physics, University of Hamburg (Germany)
References
[1] B. Weinhausen et. al., New J. Phys. 14, 085013 (2012).
[2] B. Weinhausen et. al., Phys. Rev. Lett. 112, 088102 (2014).
[3] V. Piazza et al., ACS Nano 8, 12228-12237 (2014).
[4] F. Huber et al., Curr. Opin. Cell Biol. 32, 39-47 (2015).
[5] M.P. Priebe et. al., Biophys. J. 107, 2662-2673 (2014).
[6] J.-F. Nolting et. al., Biophys. J. 107, 2693-2699 (2014).



