- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2013
- Structural biology
- How the chaperone FACT recognises histones H2A-H2B and mediates nucleosome reorganisation
How the chaperone FACT recognises histones H2A-H2B and mediates nucleosome reorganisation
The astonishing ability of chromatin to condense metres of DNA into a highly organised, regulated, but tiny structure that fits in the cell’s nucleus would not be possible without nucleosomes, the smallest repeating units of chromatin structure. Two coils of DNA wrap around a histone octamer core, which neutralises the DNA’s charges and ‘packs’ the lengthy molecule. However, nucleosomes also create a barrier to processes that require access to our genome such as DNA transcription, replication and repair. A variety of nucleosome remodelling machines and histone chaperones facilitate nucleosome dynamics by depositing or evicting histones and unwrapping the DNA. The eukaryotic FACT complex (composed of the subunits Spt16 and Pob3 in yeast) is an essential and highly conserved chaperone [1] that assists the progression of DNA and RNA polymerases and promotes the genome-wide integrity of chromatin structure, including the suppression of cryptic transcription. Genetic and biochemical assays have shown that FACT’s chaperone activity is crucially mediated by a direct interaction with the histone H2A-H2B heterodimer [2,3].
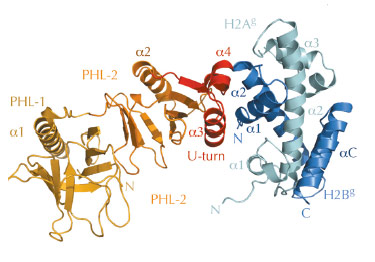
However, the structural basis for histone H2A-H2B recognition by FACT, and how this leads to nucleosome reorganisation, was unknown. Therefore, we identified biochemically the evolutionary conserved ‘chaperone’ domain of FACT, which lies within the Spt16M module. Next, we were able to solve the crystal structure of Spt16M in complex with H2A-H2B at 2.35 Å resolution (Figure 66), the first report of a globular protein in complex with these histones. Spt16M forms a tandem PHL (PH-like) module with a novel C-terminal α-helical motif, which we term the ‘U-turn’. The three-helical ‘U-turn’ clamps the α1-helix of H2B are one of the strongest histone-DNA contacts in the nucleosome. The interface between Spt16M and H2B is hydrophobic and complementary, with little difference in backbone architecture compared to the free proteins. The interaction was verified in vitro and in vivo: engineered point mutants in the interface on both the histone and the chaperone side abrogate complex formation in isothermal titration calorimetry and size-exclusion chromatography. Furthermore, consistent with the high sequence conservation of the U-turn, mutation of most U-turn residues is lethal in the budding yeast S. cerevisiae, indicating that the U-turn motif and its H2A-H2B binding function are essential.
 |
|
Fig. 66: 2.35 Å structure of the Sp16M chaperone domain (orange/red) in complex with histones H2A-H2B (light green–blue). The interface is formed by the U-turn of Spt16M (red) and the α1 helix of H2B. |
We found that electrostatic interactions contribute to the stability of the histone-chaperone complex. First, the basic N-terminal tail of H2B interacts with a conserved acidic patch on the Spt16M tandem PHL domain, and second, an unstructured stretch of ~80 mostly acidic residues adjacent to Spt16M at the C-terminus of Spt16 contributes a strong interaction with the H2A-H2B dimer. Furthermore, we found that Spt16M also recognises histones H3-H4. Similar to the structurally related chaperone Rtt106, Spt16M binds the αN helix of H3 [4].
 |
|
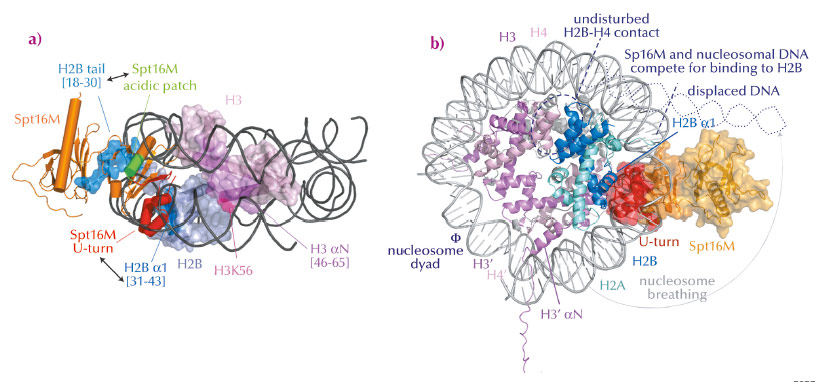
Fig. 67: Model for FACT-mediated nucleosome reorganisation. a) Overview of interactions between Spt16M and histones H2B and H3, in the nucleosomal context. b) Superposition of the Spt16MH2A-H2B complex onto the nucleosome structure. Spt16M and DNA compete for the binding site on H2B α1, therefore Spt16M binding must induce displacement of the DNA resulting in nucleosome breathing. |
In summary, we found that Spt16M makes multiple interactions with histones (Figure 67a), which has led us to suggest a model for FACT-mediated nucleosome reorganisation (Figure 67b). We propose that the chaperone gradually invades the nucleosome. First, it attaches to its binding sites on H3 αN and the H2B N-terminal tail, which are solvent-accessible. There, it would be in a good position to wait until DNA spontaneously dissociates from H2B α1, which happens about 10% of the time. By binding to H2B α1 (as we see in our crystal structure of the complex), Spt16M most likely blocks the strongest interaction surface of H2B with nucleosomal DNA and therefore out-competes and displaces ~30-50 base pairs of DNA, a process called ‘nucleosome breathing’. Such loosening of the nucleosome is presumably sufficient for DNA and RNA polymerase passage. The suggested mechanism anticipates that H2A-H2B may not need to be dissociated from the histone octamer, as suggested previously. Such interactions would thus help to preserve chromatin integrity and epigenetic memory.
Principal publication and authors
M. Hondele (a,b), T. Stuwe (b), M. Hassler (a,b), F. Halbach (c), A. Bowman (a), E.T. Zhang (b), B. Nijmeijer (b), C. Kotthoff (a), V. Rybin (b), S. Amlacher (d), E. Hurt (d) and A.G. Ladurner (a,b), Nature 499, 111–114 (2013).
(a) Adolf Butenandt Institute for Physiological Chemistry, Ludwig Maximilians University of Munich (Germany)
(b) Genome Biology Unit and Structural & Computational Biology Unit, EMBL, Heidelberg (Germany)
(c) Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Martinsried (Germany)
(d) Biochemistry Center, University of Heidelberg (Germany)
References
[1] G. Orphanides, W.H. Wu, W.S. Lane, M. Hampsey and D. Reinberg, Nature 400, 284–288 (1999).
[2] R. Belotserkovskaya, Science 301, 1090–1093 (2003).
[3] H. Xin et al., Mol. Cell 35, 365–376 (2009).
[4] D. Su et al., Nature 483, 104–107 (2012).



