- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2012
- Structural biology
- The structure of an ‘alternative’ respiratory complex I
The structure of an ‘alternative’ respiratory complex I
Mitochondria are “cellular power plants” that generate most of the cellular supply of adenosine triphosphate (ATP), which is used as a source of energy. The first reaction of the mitochondrial respiratory system is carried out by complex I. Many organisms possess alternative enzymes that can bypass or replace the complexes of the mitochondrial respiratory chain. These include the alternative NADH-quinone oxidoreductase enzymes (NDH-2), which can bypass complex I. NDH-2 enzymes catalyse the transfer of an electron from NADH via FAD to quinone.
Ndi1, an NDH-2-type enzyme, is found in the mitochondria of Saccharomyces cerevisiae (Baker’s yeast). Ndi1 can functionally replace complex I of the mitochondrial respiratory chain both in Caenorhabditis elegans (round worm) and in cultured human cells. Recently, the expression of yeast Ndi1 in Drosophila melanogaster (fruit fly) mitochondria conferred protection against toxins and the usually lethal complex I knockdown [1]. Importantly, Ndi1 has been shown to increase lifespan, independently of the effects of diet, by mitigating mitochondrial ROS production in ageing flies and limiting the accumulation of markers of oxidative damage. NDH-2 enzymes are also important targets for drug design, since their activity is crucial to the life cycles of the causative agents of malaria (Plasmodium falciparum) and tuberculosis (Mycobacterium tuberculosis).
To understand the molecular basis of the activities of NDH-2 enzymes and as a starting point for drug development, we need to know the crystal structure of Ndi1. Ndi1 is a monotopic membrane protein (possessing an integrated structure that does not cross the membrane bilayer). Monotopic membrane proteins include a significant fraction of their structure that is soluble, therefore their structural characterisation involves a delicate balance between the requirements for membrane protein stabilisation (for example, the presence of detergents) and traditional techniques applied in soluble protein structural elucidation.
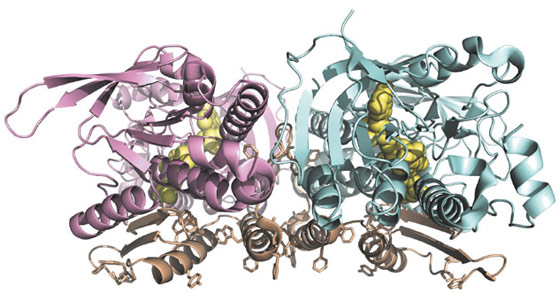
We were able to solve the crystal structure of Ndi1 to 2.7 Å resolution, with much of the diffraction data collected on beamlines ID29 and ID23-1. The structure (Figure 13) reveals three domains: the FAD-binding domain (residues 43-178 and 342-442), the NADH-binding domain (residues 179-341) and a C-terminal membrane-anchor domain (residues 443-513). The FAD- and NADH-binding domains show typical Rossmann folds and significant structural similarity to soluble FAD-dependent oxidoreductase-type enzymes. The C-terminal membrane-anchor domain is composed of a contiguous 70-residue stretch, with a three-stranded β-sheet connected to a helix-turn-helix structure. The crystals of Ndi1 contained dimeric assemblies of the protein. The membrane-anchor domains from each monomer associate in the dimer to create a contiguous apolar ridge across the centre of the complex. The base of this ridge is the part of the protein most deeply inserted into the membrane. It is comprised of the C-terminal helix of the membrane-anchor domain and presents predominantly aromatic residues to the membrane (Figure 13).
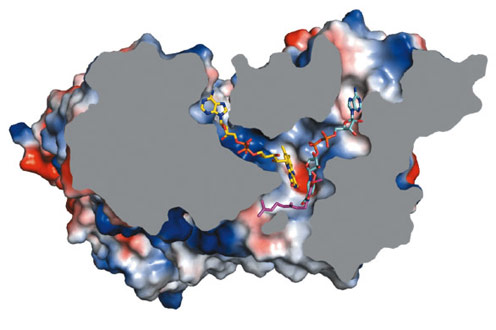
To investigate the location of the active site of Ndi1 and in particular, its substrate-binding properties, we crystallised the protein in the presence of NAD+ and UQ2 (ubiquinone-2). The resulting crystal structures show that these molecules bind in overlapping positions, stacking approximately 3.0 Å from the re-face of the FAD cofactor. This common binding site lies at the intersection of two channels in the protein structure – one leading from the matrix and the other from the membrane. Ndi1 therefore accommodates the passage of both lipid- and water-soluble substrates to a common binding site (Figure 14).
Monotopic membrane proteins lie at the bilayer’s aqueous interface and integrate into the membrane, without crossing the membrane bilayer. This type of membrane insertion is crucial in biology as it allows protein active sites of widely varying activity to engage the lipid core, without the constraints of pure α or β folds that are features of integral membrane proteins [2]. The Ndi1 structures presented here are in some ways the ultimate example of this type of adaptation. Ndi1 brings essential activity (NADH-quinone oxidoreduction) to the membrane such that the binding sites for its water-soluble (NADH) and lipid-soluble (ubiquinone) substrates overlap. This highlights Ndi1 as an example of the evolution of a membrane protein from an ancestor soluble protein, explaining how the protein adopted membrane soluble substrates.
Principal publication and authors
M. Iwata (a,b), Y. Lee (a,b,c), T. Yamashita (d,e), T. Yagi (d), S. Iwata (a,b,c,f,g,h), A.D. Cameron (a,b,c,f) and M.J. Maher (a,i), Proc. Natl. Acad. Sci. USA 109, 15247-15252 (2012).
(a) Division of Molecular Biosciences, Membrane Protein Crystallography Group, Imperial College, London (UK)
(b) Membrane Protein Laboratory, Diamond Light Source, Harwell Science and Innovation Campus, Oxfordshire (UK)
(c) Human Receptor Crystallography Project, Japan Science and Technology Agency, Kyoto (Japan)
(d) Department of Molecular and Experimental Medicine, The Scripps Research Institute, La Jolla, CA (USA)
(e) Department of Cardiovascular Physiology, Faculty of Medicine, Kagawa University (Japan)
(f) Rutherford Appleton Laboratory, Research Comple at Harwell, Didcot (UK)
(g) Department of Cell Biology, Graduate School of Medicine, Kyoto University, (Japan)
(h) Systems and Structural Biology Centre, RIKEN, Kanagawa (Japan)
(i) La Trobe Institute for Molecular Science, La Trobe University, Melbourne (Australia)
References
[1] A. Sanz et al., Proc. Natl. Acad. Sci. USA 107, 9105-9110 (2010).
[2] M.H. Bracey et al., FEBS Lett. 567, 159-165 (2004).