- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2011
- Structural biology
- The structural basis of modularity in ECF-type ABC transporters
The structural basis of modularity in ECF-type ABC transporters
Energy coupling factor (ECF) transporters form a subgroup of the ATP-binding cassette (ABC) transporter family. The members of this family are involved in the transport of a wide variety of substrates across biological membranes at the expense of ATP hydrolysis [1]. The core-architecture of ABC transporters is conserved and consists of four subunits/domains: two identical or structurally related transmembrane domains (TMDs) that form a single membrane pore through which the substrate is transported and two nucleotide binding domains (NBDs) that hydrolyse ATP. NBDs are well conserved and are considered the hallmark of the ABC transporter family. In contrast, the TMDs from different ABC transporters display a large variation in both structure and amino acid sequence.
Additional subunits/domains are present in some ABC transporters (e.g. “classical” importers found in prokaryotes rely on a water-soluble substrate binding protein domain (SBP) for the recognition of their substrates [1]).
ECF transporters are a group of ABC transporters involved in the uptake of vitamins and other micronutrients in prokaryotes [2]. These proteins are widespread among Gram-positive bacteria, many of which are pathogens (e.g. Staphylococcus aureus). ECF transporters contain two NBDs, typical for ABC transporters, and two transmembrane subunits, the EcfT subunit and S-component, which are not related in sequence. ECF transporters do not make use of SBPs for substrate binding. Instead, the integral membrane S-component binds the transported substrate with high affinity. S-components for different substrates, such as RibU (specific for riboflavin), ThiT (thiamin) and BioY (biotin), are unrelated in amino acid sequence, but intriguingly, they form active complexes with the same EcfT-NBD assembly [2,3] (the ‘energising module’). The sharing of a single energising module between many (up to 15) different S-components is unique for ECF transporters.
The structural basis of the modular S-component/energising module interactions has been unclear because of the absence of detectable sequence similarity between the different S-components. The high-resolution (2.0 Å, beamline ID23-1) crystal structure of ThiT that we determined together with the crystal structure of RibU [4] allowed us to elucidate how different S-components interact with the same energising module.
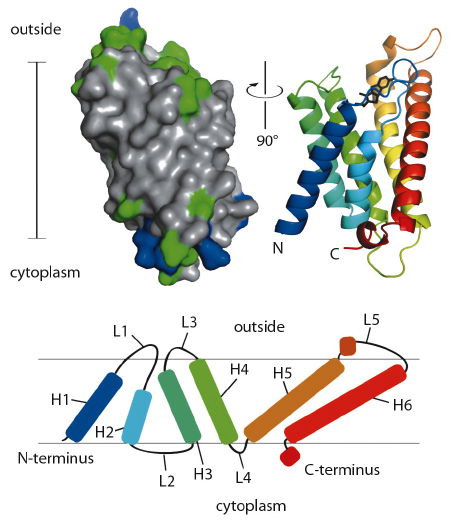
ThiT contains six hydrophobic helical segments that cross the membrane (Figure 110). A part of the loop connecting membrane helices 1 and 2 is also embedded in the lipid bilayer, and may play an important mechanistic role in the translocation of thiamin across the membrane.
 |
|
Fig. 110: Surface (left) and secondary structure cartoon representation (right) of ThiT. In the surface model, hydrophobic residues are coloured grey, hydrophilic green, positively charged blue. The cartoon is coloured from N-terminal blue to C-terminal red. The bar indicates the location of the membrane (35 Å). The topology of ThiT within the membrane is depicted at the bottom. H1-H6: helices 1-6; L1-L5: loops 1-5. |
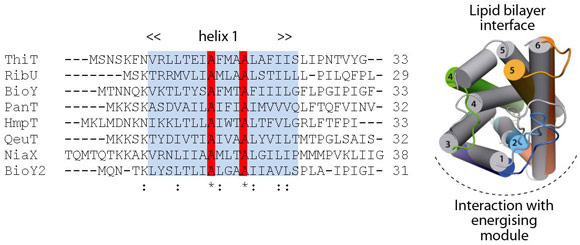
Comparison of ThiT with the structure of RibU (determined to 3.6 Å resolution) showed that the two proteins have a similar fold in spite of the lack of sequence similarity (14% identity). Membrane-spanning segments 1, 2 and 3 superimpose well, whereas segments 4 and 5 adopt different conformations. The latter are intimately involved in substrate binding, and we hypothesise that structural divergence in this region is required to bind different, chemically-unrelated substrates with high affinity (Figure 111). The structural similarity between the membrane-embedded helices 1-3 of ThiT and RibU suggests that this region is involved in the interaction with the energising module subunit EcfT. The structural alignment revealed a previously unnoticed conserved motif (AxxxA, where x is any amino acid) in transmembrane helix 1 (Figure 111). Similar motifs (e.g. GxxxG) have been shown to mediate interactions between transmembrane helices. We showed that the AxxxA motif in the S-components is important for interaction with the membrane embedded EcfT subunit: mutation of either alanine into tryptophan completely disrupted the interaction between ThiT and the energising module.
 |
|
Fig. 111: Multiple-sequence alignment of the first transmembrane helices from all S-components in L. lactis. Highlighted in red is an alanine motif that is exposed to the predicted EcfT interface in ThiT. Right: Superposition of the RibU (grey; PDB entry 3P5N, [4]) and ThiT crystal structures. |
Further structural and biochemical studies on the entire complexes will now be necessary to elucidate the complete mechanism of transport. ECF transporters are exclusively prokaryotic and numerous human pathogens are dependent on the uptake of ECF substrates for survival. Structural and mechanistic understanding of ECF transporters may therefore enable the development of new antibiotics that target these proteins.
Principal publication and authors
G.B. Erkens, R.P-A. Berntsson, F. Fulyani, M. Majsnerowska, A. Vujicic-Žagar, J. ter Beek, B. Poolman and D.J. Slotboom, Nature Structural and Molecular Biology 18, 755-60 (2011).
Groningen Biomolecular Science and Biotechnology Institute, University of Groningen (The Netherlands)
References
[1] A.L. Davidson, E. Dassa, C. Orelle and J. Chen, Microbiol. Mol. Biol. Rev. 72, 317-64, table (2008).
[2] D.A. Rodionov, P. Hebbeln, A. Eudes, J. ter Beek, I.A. Rodionova, G.B. Erkens, D.J. Slotboom, M.S. Gelfand, A.L. Osterman, A.D. Hanson and T. Eitinger, J. Bacteriol. 191, 42-51 (2009).
[3] J. Ter Beek, R.H. Duurkens, G.B. Erkens and D.J. Slotboom, J Biol. Chem. 286, 5471-5 (2011).
[4] P. Zhang, J. Wang and Y. Shi, Nature 468, 717-720 (2010).



