- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2010
- Structural biology
- Structural basis of DNA binding by human mitochondrial mTERF
Structural basis of DNA binding by human mitochondrial mTERF
Human mitochondrial DNA (mtDNA) is a circular double chain of 16.6 kDa. Its regulation is carried out by nuclear-encoded proteins, including the regulator mTERF, which binds to several strategic mtDNA sites where it controls processes such as transcription termination or replication pausing [1]. mTERF shows its highest affinity for 28 base pairs (bp) found in the tRNALeu(UUR) gene. We set out to characterise the structural basis of this interaction and crystallised two protein constructs, full-length mTERF and mTERF-ΔN (43 residues shorter than the mature protein), in complex with oligonucleotides of 15 and 12 bp, respectively, harbouring the binding site.
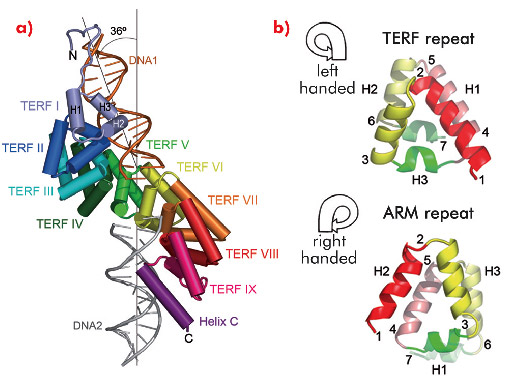
The crystal structure of mTERF (solved using diffraction data collected on ID14-4, ID23-1, ID29 and ID23-2) shows that the fold of the protein consists of a central region made up of nine structural repeats, TERF-I to IX, which correspond to tandem sequence motifs identified throughout the mTERF family, flanked by a N-terminal extension and a C-terminal α-helix, C (Figure 102a). Within each TERF repeat, roughly 35 amino acids fold into three α-helices arranged in a left-handed (zurdo in Spanish) triangular superhelix (Figure 102b, top). The nine repeats successively rotate, building up a solenoid-like structure - the zurdo domain - which further twists to the right, presenting a convex and a concave face. Previously characterised helical-repeat solenoids (Armadillo, HEAT or pumilio domains) show a right-handed helical connectivity (Figure 102b, bottom), while left-handed superhelices have been previously observed only in small helical bundles. The combination of left-handed helical connectivity with tandem propagation is thus unique to mTERF.
Two symmetrically-related 15 bp double-stranded oligonucleotides (DNA1 and DNA2) bind at positively-charged regions of the N- and C-terminal subdomains of the TERF repeats (repeats I to VI, and VIII to IX plus helix C, respectively) of a single protein molecule (Figure 102a). DNA1 and DNA2 do not contact each other and are oriented at an angle of 36º. Eighteen residues are engaged in non-specific interactions with both dsDNA backbone phosphates. In addition, Glu165, Arg169 and Arg202 at the N-terminal subdomain contact three conserved guanines; Arg387 from C-terminal helix C interacts with three bases of DNA2. Additional contacts contribute to stabilising the termini of DNA1 and DNA2. In general, the mTERF residues that contact the DNA are conserved throughout the vertebrate MTERF1 subfamily, suggesting similar modes of protein-DNA interaction for this class of protein.
 |
|
Fig. 102: a) Each of the repeats TERF-I to IX consist of three helices, H1, H2 and H3. In the crystal structure a single mTERF molecule binds two short dsDNA oligonucleotides, DNA1 (orange) and DNA2 (grey), related by ~36º. b) Left- and right-handed superhelices. From point 1 on, the TERF superhelix curves to the left while the ARM repeat shows right-handed tracing. |
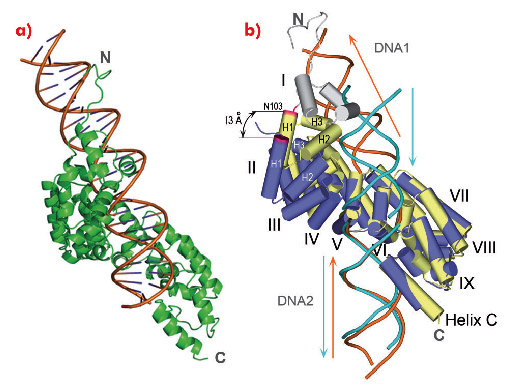
We also obtained full-length mTERF in complex with longer dsDNA oligonucleotides. However, these did not crystallise. To assess how a continuous dsDNA oligonucleotide containing both DNA1 and DNA2 could be bound by mTERF, we analysed the structure of the protein in complex with a DNA oligonucleotide of 28 bp in solution using SAXS. The model that best fitted the experimental data showed the concave surface of mTERF wrapping around the major groove of the dsDNA, which is bent (Figure 103a). The model also suggests that the C-terminal subdomain of mTERF slightly rearranges to accommodate the longer DNA. Such flexibility in mTERF is supported by the consistent, slight variation of the angle between repeats when comparing the structures of mTERF and mTERF-ΔN solenoids, whose pitch differ by 13 Å (Figure 103b).
 |
|
Fig. 103: a) The structural model showing best agreement to the experimental SAXS data. b) Comparison of mTERF (yellow) and mTERF-ΔN (blue) following a superposition of the C-terminal subdomains of their Zurdo solenoids. The maximal displacement of the N-terminal subdomain occurs at Asn103 (magenta). |
In summary, nine left-handed helical TERF repeats give rise to a twisted solenoid structure that binds a continuous bent dsDNA. The DNA binding mode observed is likely to be similar to that of mTERF binding to the mtDNA termination site in vivo. Other TERF family members are also likely to contain zurdo domains but will also have member-specific features in order to accomplish particular functions.
Principal publication and authors
N. Jiménez-Menéndez (a,b), P. Fernández-Millán (a), A. Rubio-Cosials (a), C. Arnan (a,b), J. Montoya (c), H.T. Jacobs (d), P. Bernadó (b), M. Coll (a,b), I. Usón (a,e) and M. Solà (a), Nat. Struct. Mol. Biol. 7, 891 (2010).
(a) Institute Molecular Biology Barcelona-CSIC (Spain)
(b) Institute of Research in Biomedicine (Spain)
(c) Biochemistry and Molecular and Cellular Biology, University of Zaragoza (Spain)
(d) Institute of Medical Technology and Tampere University Hospital, University of Tampere (Finland)
(e) Institució Catalana de Recerca i Estudis Avançats (Spain)
References
[1] A.K. Hyvarinen et al., Nucleic acids research 35, 6458-6474 (2007).



