- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2006
- Structural Biology
- Sensing nickel to survive the acid bath
Sensing nickel to survive the acid bath
Helicobacter pylori is a resilient bacterium that colonises the human stomach. While most organisms would be killed instantly by the acidic pH (~1), the bacterium thrives and can settle down for years. Its presence can ultimately lead to the most severe gastric diseases, including gastric ulcers, chronic gastritis and stomach cancer. Central to the bacterium’s ability to sustain such a low pH is the urease enzyme that rapidly detoxifies the environment of the bacterium by producing ammonia. The ammonia buffers the high proton content and enables H. pylori to enter the mucus layer. However, the urease enzyme requires huge amounts of nickel ions as a co-factor. This is a real dilemma for the bacterium since nickel can also be toxic at high concentrations. H. pylori has therefore developed a fine sensing mechanism that enables precise regulation of its nickel content, usage and storage.
The key element in this mechanism is the HpNikR protein (H. pylori Nickel Regulator), a transcription factor that binds specifically to DNA promoter sequences when nickel is present [1]. Although NikR proteins are present in most bacteria, their role is limited to the repression of nickel import when concentrations have reached dangerous thresholds. In H. pylori, HpNikR has multiple functions as it can repress expression of storage or import proteins as well as activating urease transcription depending on nickel concentration and/or pH value.
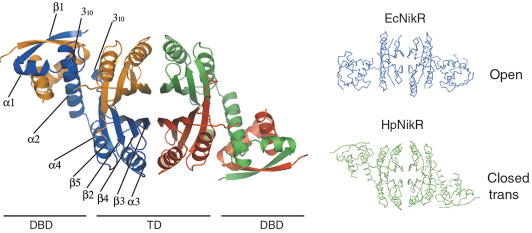
In collaboration with the Institut Pasteur, we have solved the structures of HpNikR in its apo form and in two different nickel-bound states (Ni1-HpNikR and Ni2-HpNikR) using data collected on ID14-1 and ID29. The structure of apo-HpNikR reveals that the protein has a fold similar to that of its E. coli homologue EcNikR [2] but adopts an unusual closed-trans conformation (Figure 70).
 |
|
Fig. 70: Crystal structure of apo-HpNikR and comparison with its E. coli homologue, EcNikR. |
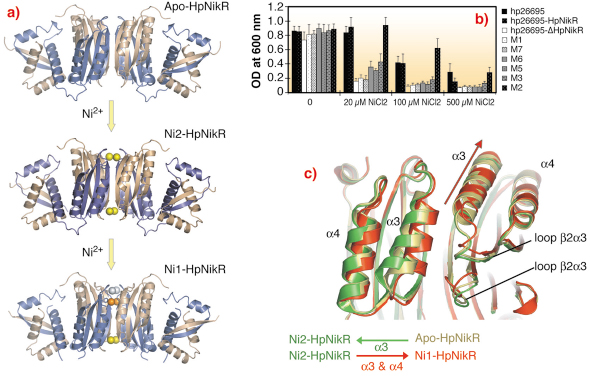
We compared the structure of apo-HpNikR with the Ni1-HpNikR and Ni2-HpNikR structures, obtained from crystals soaked in solutions containing Ni2+ and identified the route of nickel uptake within the protein (Figure 71a). Mutagenesis and in vivo complementation studies confirmed that the amino acid residues indentified by crystallography play an important role in HpNikR function. These studies therefore provide firm support for the nickel-sensing mechanism that enables the nickel response in H. pylori (Figure 71b). A close examination of the structures showed that Ni2+ entering HpNikR leads to an opening of the tetramerisation interface. The resulting movement is echoed via stacking interactions between the ![]() 3 and
3 and ![]() 4 helices and the
4 helices and the ![]() 1
1![]() 2 loops (Figure 71c). This affects other secondary structure elements involved in the DBD/TD interface. Moreover, when nickel is fully transported, the involved
2 loops (Figure 71c). This affects other secondary structure elements involved in the DBD/TD interface. Moreover, when nickel is fully transported, the involved ![]() 3 helices close down the interface (Ni1-HpNikR) and modify the position of the DBDs
3 helices close down the interface (Ni1-HpNikR) and modify the position of the DBDs
 |
|
Fig. 71: a) Ribbon representations of the crystal structures of HpNikR, in apo and nickel bound states obtained with different nickel concentrations. b) Mutational analysis of HpNikR during the acidic response. Structure based mutants were generated and cell viability was monitored upon increasing amount of nickel in the media. Clearly, mutation of residues involved in the nickel entrance in the crystal structure significantly reduce the survival of H. pylori cells. c) Ribbon representation of the movements generated by nickel incorporation in HpNikR. |
Together with previous studies, our findings provide insight into the structural basis of the nickel response in NikRs. We hypothesise that nickel transport in HpNikR involves three separate binding steps moving the Ni2+ from an external site via an intermediate site to its final high affinity site using coordinated movements of the side chains of crucial amino acids. The passage of four Ni2+ ions (one per monomer) via this route affects the dynamics of the DBDs of HpNikR which results in an active (DNA binding) form of the tetramer. Slight differences in primary sequence between HpNikR and EcNikR result in subtle changes in structure that could explain how the two proteins have different functions. For instance, occupation of a low affinity Ni2+ site present in EcNikR may not be necessary for the activation of urease transcription by HpNikR. This study firmly establishes that HpNikR is a modified, evolution driven shaped nickel sensor protein, structurally adapted to provide flexible sensor mechanisms.
References
[1] N.S Dosanjh, and S.L. Michel, Curr Opin Chem Biol 10, 123-130 (2006).
[2] E.R. Schreiter, M.D. Sintchak, Y.Guo, P.T. Chivers, R.T. Sauer, and C.L Drennan, Nat. Struct. Biol. 10, 794-799 (2003).
Principal Publication and Authors
C. Dian (a), K. Schauer (b), U. Kapp (a), S. McSweeney (a), A. Labigne, (b), and L. Terradot, (a), J. Mol. Biol. 361, 715-730 (2006).
(a) ESRF
(b) Institut Pasteur (Paris)



