- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2006
- Structural Biology
- A proton motive force driven peristaltic antibiotic pump
A proton motive force driven peristaltic antibiotic pump
Multidrug resistance is a serious threat in the fight against cancer and microbial infections. This resistance is often associated with the overproduction of membrane transport proteins that are capable of extruding chemotherapeutics, antibiotics, detergents, dyes and organic solvents from the cell. Multiple-antibiotic resistant Escherichia coli strains have been shown to overproduce two proteins: the periplasmic fusion component AcrA and the resistance-nodulation-cell division (RND) type efflux pump AcrB. In conjunction with the outer membrane channel TolC, the tripartite AcrAB-TolC system transports drugs directly into the medium, bypassing the periplasmic space and outer membrane. We solved the crystal structure of the inner-membrane component AcrB as an asymmetric trimer and postulate a novel peristaltic pump mechanism for transport of antibiotics via a tunnel with alternating access.
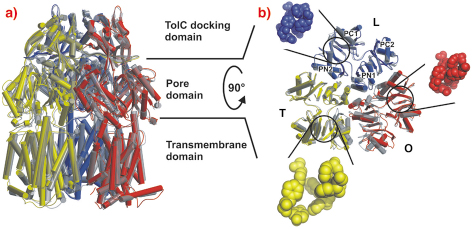
Crystals grown in a trigonal space group, assigned as R32, yielded a symmetric structure of an AcrB trimer at 3.5 Å resolution [1,2]. However, we observed merohedral twinning of the crystals in this space group, which led us to pursue the structure from different crystal forms. Eventually we solved structures of trimeric AcrB determined at 2.9 and 3.0 Å resolution in space groups (C2, P1), which do not entail three-fold symmetry of the trimer. In contrast to the symmetric structure, this asymmetric structure reveals three different monomer conformations representing consecutive states in a transport cycle, loose (L), tight (T) and open (O) (Figure 64). The structural changes in the T monomer create a hydrophobic pocket, which is not present in the other monomers. We assume that this pocket is a substrate binding pocket inside the pore domain.
 |
|
Fig. 64: Main structural differences of the AcrB monomers. The three AcrB monomers, shown in cylinder representation in blue (loose, L), yellow (tight, T) and red (open, O), are superimposed onto the symmetric AcrB trimer model [1,2], which is depicted in transparent grey. a) A side view superimposition of the entire AcrB asymmetric trimer (coloured) and symmetric AcrB (transparent grey). b) A top view superimposition of the pore domain of the asymmetric and symmetric AcrB trimer structure. The structural changes in the T monomer create a hydrophobic pocket (emphasised with yellow space-filled model) in the PN2/PC1 subdomain interface, which is closed in the L (blue) and O (red) monomers. |
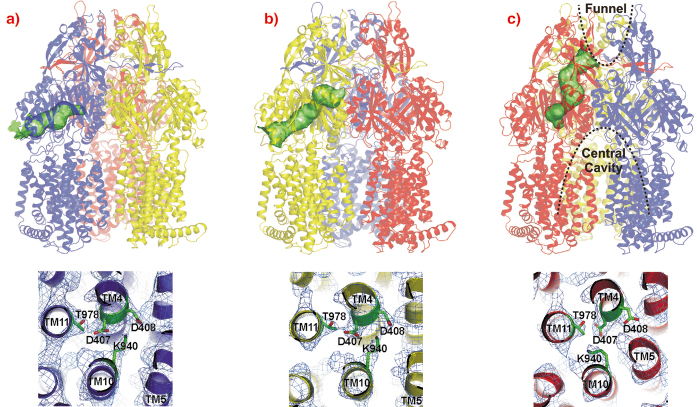
The AcrB monomers show tunnels with occlusions at different sites leading from the lateral side through the pore domains towards the funnel of the trimer and TolC (Figure 65, upper panel). From these structures, we postulate a possible peristaltic pump transport mechanism based on a functional rotation of the trimer.
 |
|
Fig. 65: Upper panel: Visualisation of the tunnels in the pore domain of the trimeric AcrB peristaltic drug efflux pump. The AcrB monomers are presented in a) blue (loose, L), b) yellow (tight, T) and c) red (open, O). The tunnels are highlighted as green surfaces in a ribbon model of the AcrB trimer. Lower panel: the putative proton translocation site with conserved residues D407, D408 (TM4) and K940 (TM10) in the three monomers is depicted as viewed from the cytoplasm with 2Fo-Fc electron density map contoured at 0.5 |
The large conformational changes observed in the periplasmic part of this protein coincide with more subtle changes in the transmembrane domain. In this domain of AcrB, TM4 and TM10 are surrounded by the other transmembrane helices of the monomer and harbour the residues K940 (TM10), D407 and D408 (TM4). As AcrB is energised by the proton-motive force, transient protonation of titratable groups within the transmembrane domain of the protein is probably the mechanism which delivers the energy required for the conformational changes described above. Indeed, we observe a prominent K940 (TM10) side chain reorientation away from D408 and towards D407 (both on TM4) in the O conformation (Figure 65, lower panel), and a bulging of TM5 towards TM4 and TM10, strengthening the hypothesis that this part of the transmembrane domain is central to proton binding and release.
In conclusion, our model merges Jardetzky’s alternate access pump mechanism with the rotating site catalysis of F1F0-ATPase and suggests a working hypothesis for the transport mechanism of RND transporters.
References
[1] S. Murakami, R. Nakashima, E. Yamashita and A. Yamaguchi, Nature 419, 587-593 (2002).
[2] K.M. Pos, A. Schiefner, M.A. Seeger and K. Diederichs FEBS Lett. 564, 333-339 (2004).
Principal Publication and Authors
M.A. Seeger (a,c), A. Schiefner (b), T. Eicher (a), F. Verrey (a), K. Diederichs (b) and K.M. Pos (a) Science 313, 1295-1298 (2006).
(a) Institute of Physiology and Zurich Centre for Integrative Human Physiology (ZIHP), University of Zurich (Switzerland)
b) Department of Biology, University of Konstanz (Germany)
(c) Institute of Microbiology, Swiss Federal Institute of Technology (ETH), Zürich (Switzerland)



