- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2005
- Structural Biology
- The Structure of a Leading Malaria Vaccine Candidate: Apical Membrane Antigen 1
The Structure of a Leading Malaria Vaccine Candidate: Apical Membrane Antigen 1
Malaria is a global health problem that severely hampers economic and social development in many third-world countries. It results in more than 2 million deaths each year and, along with HIV/AIDS and tuberculosis, is one of the three “diseases of poverty”. Malaria is caused by the unicellular parasite Plasmodium, with the two species, P. falciparum and P. vivax, being responsible for most cases of the disease in humans [1]. With the growing problem of parasite resistance to anti-malarial drugs, the search for an effective vaccine has received much attention in recent years. A number of surface proteins from Plasmodium show promise as vaccines and Apical Membrane Antigen 1 (AMA1) is one such candidate currently undergoing clinical trials. The precise biological function of AMA1 is not known but it is essential for invasion of the host liver and red blood cells by the parasite. Antibodies induced by AMA1 prevent invasion and provide the principal mechanism of protection against the disease when used in immunisations [2].
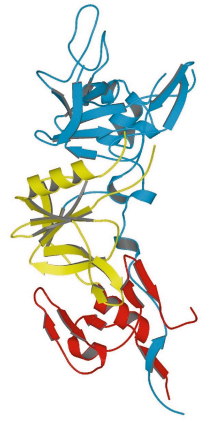
AMA1 is a type I membrane protein comprising an N-terminal ectoplasmic region of about 450 amino acids, a single transmembrane segment and a small C-terminal cytoplasmic domain. The crystal structure of the recombinant soluble ectoplasmic region from the species P. vivax has been solved by MAD using a platinum derivative (data measured on BM14 and ID14-4) and refined at 1.8 Å resolution. The three-dimensional structure of this region of AMA1 is divided into three domains (Figure 83). Although no structural similarity to known protein folds was anticipated from the amino acid sequence of the antigen, both domains I and II were found to have a fold belonging to the PAN (Plasminogen-Apple-Nematode) motif. These two domains show negligible sequence identity between each other, but their structurally equivalent regions (a total of 83 residues) nonetheless superimpose with an r.m.s. difference of 2.7 Å between the C![]() positions. Domain III does not belong to any previously identified protein fold.
positions. Domain III does not belong to any previously identified protein fold.
 |
|
Fig. 83: Structure of the ectoplasmic region of AMA1 showing domain I (blue), domain II (yellow) and domain III (red). |
Some regions of the AMA1 polypeptide chain are found as large surface-exposed insertions on the basic PAN core of domain I or domain II; these are essentially devoid of any regular secondary structure and show significant flexibility. Since AMA1 homologues are found in other members of the phylum Apicomplexa, to which Plasmodium belongs, we speculate that domains I and II derive from a gene duplication event that occurred early in the evolution of this class of unicellular organism. Little is known about general structure/function relationships of proteins bearing PAN domains, but one of their roles is the adhesion to protein or carbohydrate ligands.
Many Plasmodium surface antigens, including AMA1, have developed a high level of polymorphism (antigenic diversity) to escape the immune response of the host. In AMA1 from P. falciparum, 52 residues of the ectoplasmic region (12% of the sequence) are polymorphic. Most of this polymorphism resides in domain I, suggesting that this region plays a major role in camouflaging the parasite against the host’s immune response. Curiously, however, the distribution of polymorphic sites is highly biased to one side of the molecule, implying that antigenic variation on the other side is subject to strong functional constraints. Moreover, we have pinpointed a functionally important region of AMA1 by mapping the epitope of an invasion-inhibitory monoclonal antibody through a systematic mutation study and relating these results to the three-dimensional structure. The epitope is located at the base of a flexible 40-residue loop on domain II that is devoid of any known polymorphisms. In addition, studies on the naturally acquired immunity of individuals living in malaria-endemic regions have shown that this region also carries an immunodominant T-cell epitope. Thus, the importance of the domain II loop in the immune response and its conserved sequence between different P. falciparum isolates suggest that it might form a basis for simpler (sub-domain) vaccine constructions.
References
[1] www.who.int/topics/malaria/en/
[2] A.W. Thomas et al., Mol. Biochem. Parasitol., 13, 187-199, (1984)
Principal Publication and Authors
J.C. Pizarro (a), B. Vulliez-Le Normand (a), M.-L. Chesne-Seck (a), C.R. Collins (b), C. Withers-Martinez (b), F. Hackett (b), M.J. Blackman (b), B.W. Faber (c), E.J. Remarque (c), C.H.M. Kocken (c), A.W. Thomas (c), G.A. Bentley (a), Science 308, 408-411 (2005).
(a) Unité d’Immunologie Structurale, CNRS URA 2185, Institut Pasteur, Paris (France)
(b) Division of Parasitology, National Institute for Medical Research, London (UK)
(c) Department of Parasitology, Biomedical Primate Research Centre, Rijswijk (The Netherlands)



