- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2011
- Structural biology
- Structure of human complement iC3b revealed by 3D-electron microscopy and small-angle X-ray scattering
Structure of human complement iC3b revealed by 3D-electron microscopy and small-angle X-ray scattering
The complement system comprises over thirty soluble and membrane-associated proteins that constantly monitor the blood and tissue interstitial fluids on the lookout for pathogens, apoptotic cells and immune complexes. Central to the innate immune system, complement also plays a role in the modulation of the adaptive immune system. Activation of the central complement component 3 (C3) by the alternative pathway C3 convertase, a transient protease complex formed by the active forms of C3 (C3b) and factor B, results in an amplification cascade that leads to massive deposition of C3b onto the targeted surface, typically a bacterium surface. Surface covalent attachment of C3b upon activation involves a dramatic conformational change that exposes a highly-reactive thioester group that is sterically masked in C3. Ultimately, C3b deposition at high density contributes to the recruitment of the so-called membrane attack complex (MAC) leading to cell lysis and clearance. Protection of self-surfaces from excessive complement activation, which would result in unwanted organ damage, is achieved by fast inactivation of the C3 convertase by complement regulators. A prime route for C3 convertase inactivation is the proteolytic cleavage of C3b to iC3b, catalysed by factor I (FI) and appropriate cofactors. A final proteolytic cleavage converts iC3b into two independent molecules, C3c and C3dg, the latter of which remains attached to a membrane. Interestingly, C3b inactivation by FI-mediated proteolysis not only prevents complement consumption and collateral damage to self-cells but also generates active molecules, such as iC3b and C3dg, that can interact with leukocytes and stimulate phagocytic clearance of pathogens.
 |
|
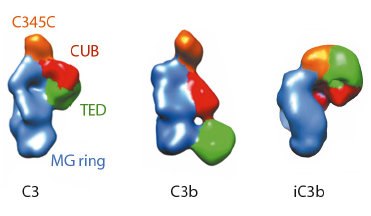
Fig. 120: Molecular architecture of iC3b as obtained by 3D-EM compared to crystal structures of C3 (PDB 2A73) and C3b (PDB 2I07) after filtering to a resolution similar to that of the structure of iC3b. For clarity, structural regions have been coloured differently. |
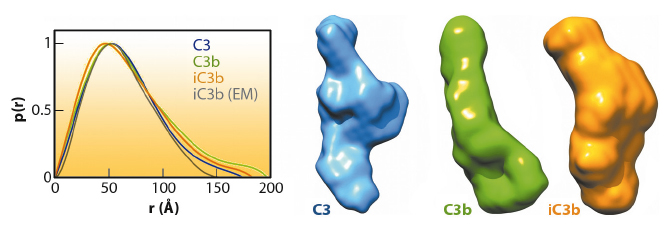
The prevailing model considers iC3b as a partially unfolded intermediate without a defined conformation [1], portrayed as two stable molecular fragments (C3c and C3dg) tethered to one another by an extended linker. This view is, however, at odds with early spectroscopic data [2] and the known role of iC3b as a specific ligand for the leukocyte complement receptors (CR), CR2, CR3 and CR4. Since a high-resolution structure of iC3b has remained elusive, we decided to piece together a medium-resolution structure using a combination of two powerful imaging techniques: single-particle electron microscopy, also known as three-dimensional electron microscopy (3D-EM), and small-angle X-ray scattering (SAXS). SAXS data were collected at the ESRF’s BioSAXS station (ID14-3) using homogenously purified proteins at concentrations ranging from 1 to 5 mg/ml. We now show using 3D-EM that the overall architecture of iC3b at low resolution (24 Å) has a distinct, defined structure (Figure 120), in contrast with previous assumptions. Powerful 3D image reconstruction techniques and in-solution shape restoration methods independently converge upon the same overall structure (Figure 121), which reveals an unforeseen conformational change. In stark contrast to the downward movement of the thioester-containing domain (TED) seen during C3 activation to C3b, in the iC3b 3D-EM structure this domain bounces back to relocate closer to its original position in C3. The macroglobulin (MG) ring at the core of iC3b however remains essentially unchanged with respect to C3b, indicating that it acts as a rigid scaffold while other domains are reoriented and translated. These features define a unique conformation that differentiates iC3b from C3 and C3b in overall shape and which has the potential for establishing distinctive protein-protein interactions with other complement components, regulators and receptors.
The iC3b structure that emerges from the 3D-EM and SAXS analysis explains an array of biochemical observations that were hard to reconcile with the more flexible ‘beads-on-a-string’ model of iC3b. In contrast to C3b, the conformational rearrangement observed in iC3b removes surfaces that had been previously identified in C3b as critical for the assembly of the C3 convertase and for recognition of FH and MCP, two complement regulators. Thus, distinctive C3b biological properties (such as the ability to form a C3 convertase) are notably absent in iC3b. Furthermore, the rearrangement exposes surface patches that are partially or totally occluded in C3b and absent in downstream fragments (C3c or C3dg), thereby creating new opportunities for iC3b to engage in interactions with receptors such as CR2, CR3 and CR4. This iC3b-specific interaction profile provides an underpinning for the biomedically relevant roles of iC3b in mediating adaptive immunological responses, stimulating B cell-mediated immunity, down-regulating inflammation and aiding the phagocytic clearance of pathogens.
Principal publication and authors
M. Alcorlo (a), R. Martínez-Barricarte (a,b), F.J. Fernández (a), C. Rodríguez-Gallego (a), A. Round (c), M.C. Vega (a), C.L. Harris (d), S. Rodríguez de Cordoba (a,b) and O. Llorca (a), Proc Natl Acad Sci USA 108, 13236–13240 (2011).
(a) CAM Complement Programme (S2010/BMD-2316), Centro de Investigaciones Biológicas (CIB-CSIC), Madrid (Spain)
(b) Centro de Investigación Biomédica en Enfermedades Raras, Madrid (Spain)
(c) European Molecular Biology Laboratory (EMBL), Grenoble (France)
(d) Cardiff University School of Medicine, Cardiff (UK)
References
[1] N. Nishida, T. Walz and T.A. Springer, Proc Natl Acad Sci USA 103, 19737-19742 (2006).
[2] D.E. Isenman, J Biol Chem 258, 4238-4244 (1983).