- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2010
- Structural biology
- Binding of the signal peptide to the signal recognition particle
Binding of the signal peptide to the signal recognition particle
Many proteins are inserted into the cell membrane or secreted from the cell. In 1972, César Milstein, working at the MRC Laboratory of Molecular Biology, discovered that the immunoglobulin light chain is made as a longer precursor; a short sequence at the beginning of the protein is cleaved off when it is secreted. He suspected that this short sequence, now known as the signal peptide, might direct proteins to outside the cell [1]. Signal sequences are divisible into a positively charged n-region, an h-region commonly containing 8–20 hydrophobic residues and a polar c-region, but all regions lack sequence conservation [2]. When the signal sequences (or signal peptide) of secreted proteins emerge from the ribosome they are bound by a signal recognition particle (SRP). SRP directs the ribosome to the endoplasmic reticulum membrane in eukaryotic cells or plasma membrane in prokaryotic cells where the ribosome docks with a protein-conducting channel (translocon). Synthesised proteins are then inserted into the membrane or secreted through a translocon co-translationally [3-5]. It was not known how the signal peptide binding site of SRP54 can recognise highly diverse signal sequences. We have now crystallised SRP54 bound to a signal peptide showing molecular details of recognition.
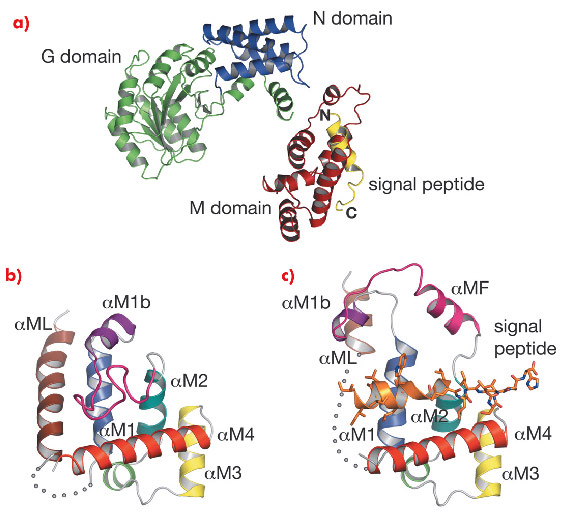
Signal recognition particles are found in all three domains of life (Eubacteria, Archaea and Eukarya) but their structures differ greatly with only SRP54 (or its homologue Ffh) as the universally conserved protein component [4-5]. SRP54 consists of three domains: the N-terminal four-helix bundle (N domain) and a Ras-like GTPase domain (G domain), which together form the NG domain, and the carboxy-terminal methionine-rich M domain. SRP54 associates with SRP RNA and signal peptide through its M domain. SRP interacts with the ribosome near the peptide-exit site mainly through the N-domain [5] and captures the signal sequence as soon as it emerges from the ribosome. The SRP-ribosome-nascent chain complex (SRP-RNC complex) is then directed to a translocon via a GTP-dependent interaction between the NG domains of SRP54 and a membrane-associated SRP receptor. No structure has been reported that exemplifies SRP54 binding of any signal sequence because of the difficulties in producing a complex of SRP54 and hydrophobic signal peptides. We produced SRP54 fused with a signal peptide via a linker peptide. This dimeric form of SRP54* crystallised in space group P41212 with one monomer per asymmetric unit. Data were collected at beamlines ID14-1 and ID14-4. The structure was solved by the MAD method using a methylmercury derivative of a single cysteine mutant (N177C) and refined to an Rfree of 32.3% at 3.5 Å resolution. An unbiased electron density map calculated by combining the Hg MAD phases with molecular replacement phases from a homologous NG domain (PDB code: 1J8M) followed by solvent flattening showed clear density of the signal peptide (PDB code: 3KL4).
 |
|
Fig. 111: a) Structure of Sulfolobus solfataricus SRP54–signal peptide fusion protein. b) The M domain of free SRP54 (PDB code: 1QZX). c) The M domain of SRP54* in complex with a signal peptide (residues Gly 449B–His 468B). The poorly ordered region (residues 308–326) is represented by a dotted curve. |
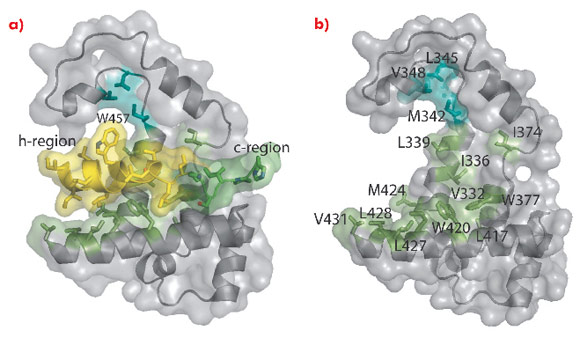
In the M domain of SRP54* helix αM4, and helices αM1 and αM2 oriented perpendicular to αM4, form a hydrophobic groove where the signal peptide in the α-helical conformation is accommodated (Figure 111). The indole rings of Trp 377 and Trp 420 cluster together creating a bulge in the groove surface. Eight residues (Ile 450–Trp 457) in the h-region of the signal peptide form an a helix which extends to Leu 461 with a bend at Gly 458 that complements the Trp bulge in the groove floor (Figure 112). The signal peptide helix and αM4 interact by the most commonly occurring 4-4 ‘ridges-into-grooves’ helix packing, with a crossing angle of –40° in the Chothia notation [6]. This crossing angle allows the observed extensive interaction between the h-region of the signal peptide and aM4 of the M domain. In addition, the Leu and Met side chains, a conserved hallmark of the M domain, can adopt different rotamers allowing signal peptides with diverse sequences to bind.
The structure of SRP54* has provided a first structural insight into signal peptide binding to SRP. It would be very interesting to crystallise SRP54 with different signal sequences to see how residues in the signal peptide binding groove adopt different conformation to optimise interactions.
Principal publication and authors
C.Y. Janda (a,b), J. Li (a), C. Oubridge (a), H. Hernandez (c,d), C.V. Robinson (c,d) and K. Nagai (a), Nature 465, 507–510 (2010).
(a) MRC Laboratory of Molecular Biology, Cambridge (UK)
(b) Current address: Howard Hughes Medical Institute, Stanford University School of Medicine (USA)
(c) University of Cambridge Chemical Laboratories, Cambridge (UK)
(d) Current address: Department of Chemistry, University of Oxford (UK)
References
[1] C. Milstein, G.G. Brownlee, T.M. Harrison and M.B. Mathews, Nature New Biol. 239, 117-120 (1972).
[2] L.M. Gierasch, Biochemistry 28, 923–930 (1989).
[3] P.F. Egea, R.M. Stroud and P. Walter, Curr. Opin. Struct. Biol. 15, 213–220 (2005).
[4] J.A. Doudna and R.T. Batey, Annu. Rev. Biochem. 73, 539–557 (2004).
[5] C. Wild, M. Halic, I. Sinning and R. Beckmann, Nature Struct. Mol. Biol. 11, 1049-1053 (2004).
[6] C. Chothia, M. Levitt and D. Richardson, J. Mol. Biol. 145, 215-250 (1981).