- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2009
- Structural biology
- Studies of UvrA2 provide insight into DNA binding and damage recognition
Studies of UvrA2 provide insight into DNA binding and damage recognition
DNA damage is a common occurrence that compromises the functional integrity of DNA. DNA repair pathways are ubiquitous and the principles of damage recognition are conserved from bacteria to humans. Nucleotide excision repair is the primary pathway for repair of the structurally diverse lesions caused by ultra-violet light and involves the recognition and removal of damaged DNA by a dual-incision event. In prokaryotes, the Uvr proteins carry out the excision and have been the focus of many studies for over 20 years. The proteins UvrA and UvrB are responsible for the ATP-dependent recognition of DNA damage whilst UvrC is required for the subsequent incision events. Despite extensive biochemical and genetic studies, it is still not well understood how nucleotide excision repair is capable of recognising such a wide variety of lesions, but as structural information on the UvrABC proteins is becoming available and through in depth biochemical analysis of the repair process, a picture is starting to emerge.
UvrA is responsible for initial DNA damage recognition and subsequently acts as a molecular matchmaker by efficiently promoting the stable assembly of UvrB onto damaged DNA sites. A more comprehensive description of the molecular mechanisms of UvrA’s action would therefore constitute an important contribution to our understanding of the initial steps in bacterial nucleotide excision repair. In this work, we have undertaken a structural and biochemical study of a UvrA protein (drUvrA2) from the radiation-resistant bacterium Deinococcus radiodurans. Unlike E. coli, D. radiodurans’ genome encodes two UvrA proteins, drUvrA1 and drUvrA2. drUvrA1 represents the full-length UvrA protein, which is present in all bacteria and has recently been classified as a Class I UvrA. Recently, the first three-dimensional structure of such a Class I UvrA from Bacillus stearothermophilus (bstUvrA) was reported [1]. drUvrA2, a Class II UvrA, displays a high sequence identity with drUvrA1, E. coli UvrA and bstUvrA, but is missing the UvrB binding domain recently identified in bstUvrA. Although the exact roles and substrates of Class II UvrAs remain elusive, the high level of conservation suggests that the molecular mechanisms involved in substrate recognition are most likely similar to those used by Class I UvrAs.
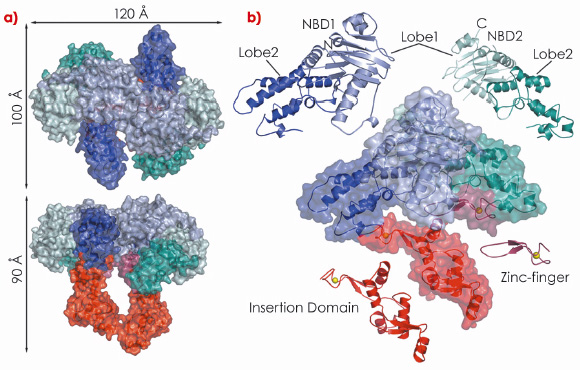
In our study, we show that drUvrA2 shares many of the biochemical (ATPase activity, nucleotide-dependent dimerisation, preferential binding to damaged DNA) and structural features observed for UvrAs. We have solved and refined the structure of ADP-bound drUvrA2 in two crystal forms to resolutions of 2.3 Å (C2; 3 mol/asu) and 3.0 Å (C222; 2 mol/asu) respectively, providing us with a view of five distinct monomeric and three dimeric conformational states. drUvrA2 assembles as a head-to-head dimer in which each monomer consists of two tandemly arranged ATPase binding cassettes (Figure 121). As expected from primary sequence analysis, our structure of drUvrA2 reveals that the UvrB-interacting domain is missing along with its associated Zn-binding site (two CXXC motifs). As a result, drUvrA2 possesses only two of the three Zn-binding sites found in the classical Class I UvrAs. As in the case of bstUvrA, a large insertion domain was accommodated in the amino-terminal nucleotide binding domain (NBD-I) of drUvrA2, while a more classical zinc-finger is inserted into the carboxy-terminal NBD-II.
 |
|
Fig. 121: Crystal structure of D. radiodurans UvrA2. Surface and ribbon representations of dimeric (a) and monomeric drUvrA2 (b). The domains are coloured as follows: NBD1 (blue), NBD2 (cyan), insertion domain (red) and zinc-finger (raspberry). Zinc ions are illustrated as yellow spheres. |
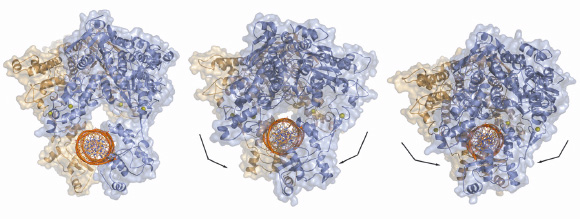
Our studies have also shown that drUvrA2 alone can recognise and bind preferentially to damaged bases within DNA oligonucleotides. This work was complemented by a mutational study of drUvrA2 that showed that two regions of UvrAs are essential for DNA binding: a positively charged groove formed on the concave side of the core ABC domains and the newly identified insertion domains. Taken together our structural, biochemical and mutational analysis of drUvrA2 provides the first detailed description of a class II UvrA and also contributes to an extended model for how UvrAs might interact with DNA (Figure 122), which has implications for a fuller understanding of the mechanisms involved in the initial steps of nucleotide excision repair.
 |
|
Fig. 122: Models of dsDNA binding to symmetrical dimers assembled from the three different molecules of drUvrA2 refined in C2 space group. |
Reference
[1] D. Pakotiprapha, Y. Inuzuka, B.R. Bowman, G.F. Moolenaar, N. Goosen, D. Jeruzalmi and G.L. Verdine, Mol. Cell 29, 122 (2008).
Principal publication and authors
J. Timmins (a), E. Gordon (a), S. Caria (a), G. Leonard (a), S. Acajjaoui (a), M.-S. Kuo (b), V. Monchois (b) and S. McSweeney (a), Structure 17, 547–558 (2009).
(a) ESRF
(b) PX-Therapeutics, Grenoble (France)



