- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2006
- X-ray Absorption and Magnetic Scattering
- Bacterial S-layers as template for the formation of Pd nanoclusters
Bacterial S-layers as template for the formation of Pd nanoclusters
Nanocatalysts made from palladium and other noble metals promise to accelerate chemical reactions even at low temperatures. Critical for the efficiency of these catalysts is a perfect control of their cluster-size. By using the surface protein layer (so-called S-layer) of a bacterium, particles with a uniform size distribution of 50 to 80 Pd atoms can be generated. For the first time, the bonding sites between the metal and the S-layer protein have been characterised. Hereby, the prerequisite is given to manipulate this protein by genetic engineering enabling the design of materials with new optic, magnetic and catalytic properties.
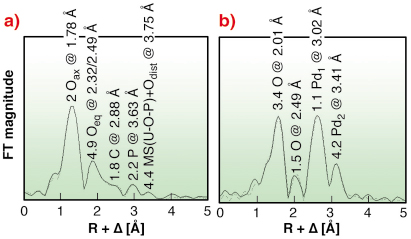
To produce nanoparticles of the noble metal palladium, a team of biologists from the Forschungszentrum Dresden-Rossendorf (FZD)/Germany use the S-layer of Bacillus sphaericus strain JG-A12. The biologists discovered this bacterium in 1997 from a uranium mining waste pile and have since then cultivated it in the laboratory. The bacterium survives in the extreme environment because it is able to tolerate high amounts of heavy metals including uranium [1,2]. One part of its survival strategy seems to be the formation of a protective outer layer, the S-layer, which exhibits a high metal-binding capacity, thereby preventing the cellular uptake of heavy metals. The S-layer protein of this bacterium is regularly structured with pores of identical size on the nanometre scale. The uranium uptake mechanism was investigated by EXAFS performed at the Rossendorf Beamline (BM20). The measurements revealed that the purified S-layer proteins of this bacterium coordinate uranium through phosphate groups of phosphorylated serine and threonine in a monodentate mode as well as through carboxyl groups of aspartic and glutamic acids in a bidentate mode (Figure 105a) [2].
 |
|
Fig. 105: a) U LIII-edge EXAFS of U(VI) complexes, and b) Pd K-edge EXAFS of Pd(II) complexes, formed by S-layers of B. sphaericus. |
The heavy metal binding capacity of the S-layer led to the idea to use it as template for the formation of Pd nanoclusters. Within the pores of the S-layer, isolated from vegetative bacteria, Pd(II) solutions are reduced to metallic palladium by the use of hydrogen. This procedure forms nanoclusters of metallic palladium, with a uniform particle size of 50 to 80 Pd atoms, which are regularly arranged on the surface layer (Figure 106).
 |
|
Fig. 106: Schematic view of Pd nanoclusters located at the S-layer protein. |
In order to investigate the metal-protein interactions and their impact on the secondary structure, a solution of Pd(II) ions has been adsorbed on the S-layer matrix. The combination of Fourier transform infrared spectroscopy and EXAFS spectroscopy revealed that the surface Pd(II) is predominantly coordinated by aspartic and glutamic residues through four nitrogen and oxygen atoms at a distance of 2.01 Å (Figure 105b). In contrast to U(VI), that binds to carboxyl and phosphate groups, Pd(II) binds exclusively to the carboxyl groups of the S-layer. The topology of nitrogen- and carboxyl-bearing side chains appears to mediate the binding of heavy metals to aspartic and glutamic acids. These side chains are thus targets for the design of engineered S-layer based nanoclusters.
Because the metal stabilises the protein and vice versa, the S-layer stays intact at higher temperatures or even in an acidic environment. By modifying the distribution of functional groups and the ternary structure of the proteins, the Pd cluster size can be modified and a uniform size distribution can be achieved. A few laboratories are already producing and testing this new technology.
References
[1] C. Hennig, P.J. Panak, T. Reich, A. Rossberg, J. Raff, S. Selenska-Pobell, W. Matz, J.J. Bucher, G. Bernhard, H. Nitsche, Radiochim. Acta 89, 625-631 (2001).
[2] M.L. Merroun, J. Raff, A. Rossberg, C. Hennig, T. Reich, S. Selenska-Pobell, Appl. Environ. Microbiol. 71, 5532–5543 (2005).
Principal Publications and Authors
K. Fahmy (a), M. Merroun (b), K. Pollmann (b), J. Raff (b), O. Savchuk (a), C. Hennig (b,c), S. Selenska-Pobell (b), Biophysical J. 91, 996-1007 (2006).
(a) Institute of Nuclear and Hadron Physics, Division of Biophysics, Forschungszentrum Dresden-Rossendorf (Germany)
(b) Institute of Radiochemistry, Forschungszentrum Dresden-Rossendorf (Germany)
(c) Rossendorf Beamline, ESRF (France)



