- Home
- Users & Science
- Find a beamline
- ID28 - Inelastic Scattering II
- User Guide
- Handling the instrument
Handling the instrument
Starting Experiment
Motor status, moving motors, and counting
Position of the alignment detector and S(Q) detector
Make sure that the premonochromator and the mirror are optimised
Put the horizontal spectrometer arm to the correct scattering angle
Put the monochromator to the starting temperature
Doing a temperature scan
Stopping a scan
Running a Macro
Plotting data file (in CPLOT)
THINGS NOT TO DO
SAVE DATA OF YOUR EXPERIMENT!!!!!!!!!!!!!
Starting Experiment
Your local contact will set the name of the experiment directory and the name of the experimental file. This is usually only one file, where each scan has an increasing number within this datafile. Example: filename exp102, scan #S 5
Motor status, moving motors, and counting
fourc> wa (shows all the motor positions)
fourc> wm (motor name) (shows the position of the particular motor)
fourc> ct (n) (counts for n seconds)
fourc> umv (motor name) (target value)
fourc> umvr (motor name) (change of actual position) (relative movement with respect to actual value, motor moves back to initial position afterwards) Example: umvr tth 0.5
fourc> dscan (motor name) (-rel. change) (+rel. change) (number of points) (counting time)
(scan around actual position, motor remains at end position) Example: dscan say 1 -1 20 1
fourc> ascan (motor name) (start value) (end value) (number of points) (counting time)
(scan from actual position to end value) Example: ascan say 2 5 30 1
For th and tth, the scans should be done from large to small angles, in order to avoid the backlash movement for each point. For all the other motors the backlash is activated for movements towards negative values. Be careful when moving these motors over large distances. Pieces of equipment or the sample chamber might be in the way and might get damaged!
Position of the alignment detector and S(Q) detector
Detector newdet in direct beam at tth = 0.666° (07.3.2003)
This value may vary. For the exact position ask your local contact.
S(Q) detector (detsq) at tth = :__ and zdetsq1 = 36.65
(zdetsq1 out = 13.3)
Make sure that the premonochromator and the mirror are optimised
Before starting an inelastic scan, you have to make sure that the premono angle mono is optimised with respect to the backscattering monochromator (correct value of imirr/pmoni). Furthermore, the mirror angle roty might need an optimisation so that the focussed beam is properly centred through the ione-slit unit (correct value of ione/imirr or ione/izero). To scan these motors for optimisation the monochk and mirrorchk routines have to be deactivated, if there were active, or alternatively be set up properly.
Set-up of mono check routine
fourc> offmonochk fourc> monochksetup
A menu appears on the screen:
MONOCHK SETUP
mono monitor counter mnemonic: imirr high threshold: 80 type: integrating machine current counter mnemonic: pmoni low threshold: 50 type: integrating sleep time after beam is back: 600 lineup mono –0.002 0.002 20 3 tweak value: 0.0003 line up at: CEN plot filter 1 pre-scan magnitude magnitude after beam loss: 0.01 pre-scan magnitude intervals after beam loss: 40
The lowest level of check is associated with the presence of the x-ray beam on the premono. If the beam is gone, the spectrum stops automatically, detects when the beam is back, and waits the predefined sleep time after the beam is back. It then performs a pre-scan of the premono, followed by the line-up scan, after which the intensity ratio imirr/pmoni should be optimised.
If during a scan the ratio imirr/pmoni drops below 80% of the initially optimised value, the mono performs a small correction to both sides in order to improve the imirr/pmoni ratio.
Set-up of mirror check routine
fourc> offmirrorchk
fourc> mirrorchksetup
A menu appears on the screen:
MIRRORCHECK SETUP
mirror monitor mnemonic: i10 threshold: 80 type: analog roty scan counter mnemonic: ione lineup roty –0.01 0.01 40 4 roty tweak value: -0.0015 line up at: CEN
This check routine makes sure that the beam is always properly steered through the ione-slit unit. This unit accommodates the last set of slits before the sample. For small samples and high pressure experiments, the vertical slit (svg) is put to 100 mm typically, and during an IXS scan, the mirror needs to be adjusted from time to time. If the intensity ratio ione/imirr (or ione/izero) drops below 80% of the initially optimised value, a small angular correction (defined by the tweak value) is performed.
If the beam is lost, first the monocheck routine is executed, then the mirrorcheck routine. It consists of a line-up scan, after which the mirror is positioned at the maxiumum of the ione/imirr signal.
Activation of the check routines for the premono and the mirror
fourc> plotselect imirr fourc> dscan mono –0.002 0.002 20 3 fourc> umv mono MAX
You have to move it to its position several times until mono really reaches this position. Before setting the monochk routine you have to continue with the optimisation of the mirror (roty) (otherwise the mirror optimisation will be hindered by the running monochk).
fourc> plotselect ione
fourc> dscan roty –0.01 0.01 40 3
fourc> umv roty MAX
You have to check whether the mirror has really reached the optimum positions. If this is not the case, you have to try to optimise it by hand, using small corrections of the order of the defined tweak value.
Now after the complete optimisation set first the monochk routine:
fourc> onmonochk 3
This activates the premonochromator (mono) control, as well as the beam control. Then set the mirrorchk routine:
fourc> onmirrorchk 3
This activates the mirror control.
Put the horizontal spectrometer arm to the correct scattering angle
fourc> umv tth 20
the units are in degrees. Look at the table at the end of the document for the conversion angle → Q(nm-1).
Put the monochromator to the starting temperature
Put a set point for the monochromator temperature by giving the values in degree °C to the pseudo-motor monot as a normal motor positioning (umv monot your_set_point) e.g.:
fourc> umv monot 22.00
You have to wait typically 5 to 10 minutes until the setpoint is reached. You can also give the values in meV using the pseudo-motor deltae, for doing this you have to enter the zero-energy temperature setpoint:
fourc> T0=actual_average_zero_Temp
and the wavelength you're using (e.g. for the (999) reflection. A complete list of the reflection orders and the corresponding wavelengths are given at the end of the document):
fourc> LAMBDA=0.6968 (UPPER CASE for the variables, it is case sensitive)
otherwise your deltae value (see below) does not make any sense.
Doing a temperature scan
| fourc> |
ascan monot init_val final_val number_of_step integration time |
Alternatively you can use the deltae motor:
| fourc> |
ascan deltae init_val final_val number_of_step integration time |
Example: scan from -80 to 80 meV, with 0.5 meV step, 1mn int. time, at the (999) reflection (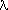 = 0.6968 Å), with T0=21.71, looks like:
= 0.6968 Å), with T0=21.71, looks like:
| fourc> | T0=21.71 |
| fourc> | LAMBDA=0.69680026 |
| (just once, after you have selected the mono-reflection and determined the elastic temperature) | |
| fourc> | ascan monot 23.6 20.08 320 60 |
or
| fourc> | ascan deltae -80 80 320 60 |
You can also type:
| fourc> | ascan monot T0-1.76 T0+1.76 320 60 |
Stopping a scan
fourc> [CTRL-C]
Scanning or counting will be stopped. If a dscan was running, the motor will return to its initial position.
Running a Macro
This is frequently utilised, if a sequence of scans shall be performed, once the measurement strategy has been defined.
1) Edit/modify the macro with the editor you like (vi, emacs, nedit etc.) in the working directory. The name of the macro (first line) must be the one of the macro file (example: name of the macro macfilename, name of the file macfilename.mac).
Example:
def nbse3 '
{
plotselect deta1
premaon
T0=21.71
umv as1vg 60
umv as1hg 20
#################################
# Analyzer #2 @ (8.35,0,0)
umv monot T0+0.4
umv wheel 315
ubr 8.35 0 0
umv wheel 0
#
ascan monot T0+0.4 T0-0.4 100 95
umv monot T0+0.4
sleep(600)
ascan monot T0+0.4 T0-0.4 100 85
################################
umv wheel 315
umv monot T0
}
'
2) Macro compilation:
fourc> qdo macfilename.mac
You can check the content of the compiled macro by
fourc> prdef macfilename
3) Macro execution:
fourc> macfilename
Plotting data file (in CPLOT)
This operation generates the standard output for the log book, and it is highly recommended. It makes it a lot easier to detect any potential bug in the spectrum and identify the problem, if the key characteristics are printed out.
Start Cplot in the fourth window of the X-Terminal: cplot
Assume the Cplot-macro is called cpfilename.do
a) Output on screen:
PLOT-> do cpfilename.do 010 70 6.25
b) Output on printer:
PLOT-> do cpfilenamep.do 010 70 6.25
"010" is scan number in the datafile, "70" counting time per point, "6.25" is Q-transfer in nm-1.
THINGS NOT TO DO
1) Move the spectrometer arm to small Q-values, where either analysers #1, #6, #2 or the S(Q) detector are close or in the direct beam (or an intense Bragg reflection).
If this is necessary put by all means the absorber wheel in:
fourc> umv wheel 315 (270)
but not less than 270, this will saturate the detector.
2) Move the sample angle theta (th) to high angles or spectrometer arm to large scattering angles (tth), since you might hit into the sample vacuum can. Visual inspection is indispensable!!!!
Be as well careful when moving sample tilt angles (phi, chi) by larger amount; there is very little space available around the sample position! The sample chamber may run into surrounding equipment or parts of the instrument (detector chamber)!
For any problems phone your local contact, or the Experimental Hall Operators (25-25).
SAVE DATA OF YOUR EXPERIMENT!!!!!!!!!!!!!
Put your data on your user account (1-month lifetime). Connect on NICE (Solaris or Linux system) using the name of your experiment (typically HSXXX), and your password (initial of the name of the first applicant followed by the family name e.g. experiment HS123, first applicant Mr. H. Gbhkds:
telnet (or ftp) nice
login: hs123 (this is case sensitive)
password: gbhs123.
Then transfer the data home using ftp out or telnet out (literally type ftp out or telnet out then follow the instructions).
For any computing problem phone to the 24-24 (computing hot line, 7 a.m.-8 p.m.).



