- Home
- News
- General News
- How mixing AlphaFold...
How mixing AlphaFold with synchrotron and cryo-EM data unveiled proteins that trick bacteria
10-03-2022
A team led by Karolinska Institutet (Sweden) has combined Artificial Intelligence (AI) with structural biology at the ESRF and SciLifeLab to determine structures of two similar proteins that prevent bacterial infection in the urinary tract and the gastrointestinal system. Their results are published in Nature Structural & Molecular Biology.
Urinary tract infection affects about 150 million people per year and is the most common form of bacterial infection in women1. Filaments of a molecule called uromodulin (UMOD), the most abundant protein in the urine, play a key role in counteracting bacterial invaders.
The surface of the urinary tract is not protected by mucus, which makes it vulnerable to bacteria. However, the body produces UMOD as a soluble defense. UMOD carries a special sugar chain that is not modified like others and remains of high-mannose type. This chain acts as a decoy by mimicking the ones to which bacteria would bind to initiate the infection in the urinary path3. As a result, bacteria bind to the high-mannose sugar chain of UMOD instead of the urinary tract, and are eventually wrapped by the UMOD protein filaments and removed through the urine.
During recent years, evidence has been accumulating that glycoprotein 2 (GP2), a molecule produced in the pancreas and the intestine, plays a similar role as UMOD in counteracting bacterial infection at the level of the gastrointestinal system. Because the GP2 and UMOD genes originated by duplication of a single ancestral gene, the two proteins share a similar architecture and ability to form filaments. However, until today it was not clear if the molecular basis of their antibacterial action was also conserved.
“Based on these considerations and our previous work on UMOD and related molecules, it was a natural choice for us to study both proteins in parallel” explains Luca Jovine, professor at Karolinska Institutet and corresponding author of the paper. “In particular, we wanted to determine which sugar chain of GP2 corresponds to the high-mannose chain of UMOD and understand how the proteins manage to keep these sugars ‘special’ so that they can act as a bait for bacteria”, he adds.
Obtaining structural information on either protein has proved a difficult task. UMOD forms long filaments that can be studied at moderate resolution by cryo-electron microscopy (cryo-EM), but the part of the molecule that is involved in binding the bacteria is particularly flexible and very different in sequence from any known protein structure. In parallel, crystals of the corresponding part of GP2 were measured at different ESRF beamlines (ID23-1, ID30B and ID30A-3), but — although different crystal forms were ultimately obtained — it was initially not possible to obtain good enough experimental phases to determine the structure.
Artificial Intelligence comes in the picture
It was at this point that the scientists began a collaboration with DeepMind and the team behind AlphaFold — a machine learning program that attracted significant attention from the structural biology community by generating highly accurate protein structure predictions during the 14th iteration of CASP (Critical Assessment of Techniques for Protein Structure Prediction). “The structure of what we identified as the decoy module of UMOD and GP2 was expected not to look like any already known protein”, says Jovine. “It turned out that, although AlphaFold had never ‘seen’ such a fold before, its predictions were extremely good. This allowed us to quickly make sense of the GP2 X-ray data, as well as to fit low resolution cryo-EM maps of UMOD — both by itself and in complex with FimH, the protein that bacteria use to bind high-mannose sugar chains. Whilst AlphaFold does not currently predict the structure of sugars, by combining it with experimental data we got the best of both worlds”, he explains.
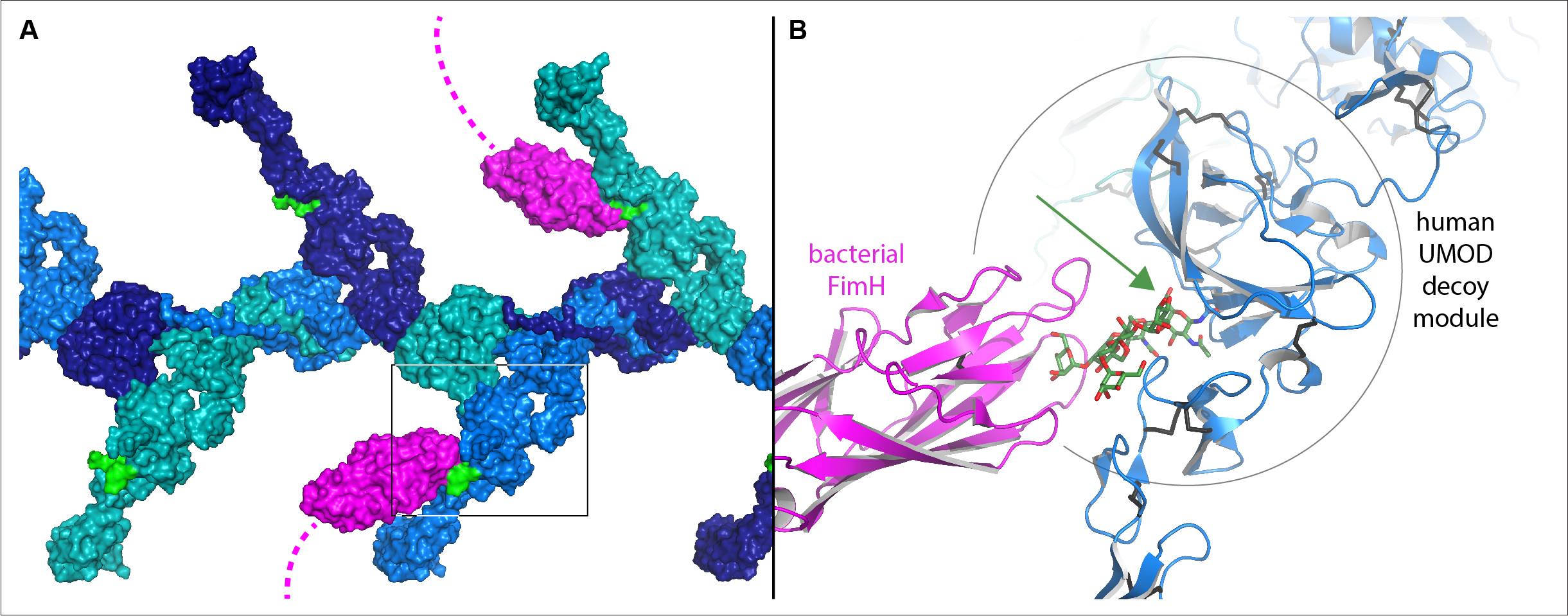 |
|
Model of the human UMOD/FimH interaction, assembled by combining the structural information described in the study. A, Surface representation of a UMOD filament segment, with subunits colored in alternating shades of blue. The high-mannose sugar chain (green) of the decoy modules of UMOD, which branch out from the filament core, is bound by the FimH sugar-binding domain at the tip of bacterial adhesive filaments (magenta). B, Detail of the region boxed in (A), showing how the sugar chain recognized by FimH protrudes from the crevice between the two parts of the UMOD decoy module (green arrow). Credits: Stsiapanava A. et al., Nature Structural & Molecular Biology, in press (DOI: 10.1038/s41594-022-00729-3). |
“There is a big ongoing discussion about the impact of AI on structural biology. We were amazed by the performance of AlphaFold, which has been a crucial tool to obtain these results. In our view, there is no doubt that machine learning is already fueling the next leap in structural biology and positioning itself as an ideal companion to X-ray crystallography, cryo-EM and bio-SAXS studies”, explains Daniele de Sanctis, scientist at the ESRF and co-author of the paper.
The results, obtained in collaboration with Nanyang Technological University (Singapore), SciLifeLab (Stockholm) and Lille University, showed that the functionally crucial decoy module of UMOD and GP2 consists of two parts separated by a crevice. Although the sugar chains that bind bacteria are attached to different regions of the GP2 and UMOD sequence, the 3D structures reveal that, in both cases, their base is positioned within the crevice. This explains how these specific sugar chains are shielded from modification and can thus be presented as hooks for fishing pathogenic bacteria.
Could these findings help to treat diseases? “Because the GP2 decoy module can be expressed at relatively high levels in a recombinant form that binds bacteria, it could, in principle, be explored as a decoy alternative to conventional antibiotic treatments. At the same time, our structural work explains many human UMOD mutations associated with kidney diseases, as well as provides information on a UMOD protein region whose interaction with filtered light chains is implicated in cast nephropathy," concludes Jovine.
Reference:
Top image: Surface representation of a UMOD filament segment, with subunits colored in alternating shades of blue. The high-mannose sugar chain (green) of the decoy modules of UMOD, which branch out from the filament core, is bound by the FimH sugar-binding domain at the tip of bacterial adhesive filaments (magenta). Credits



