- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2009
- Structural biology
- The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit
The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit
Every year influenza virus causes seasonal epidemics that affect 5-10% of the world population and kill up to 500,000 people. In nature, the virus primarily infects aquatic birds which can further infect domestic chickens and pigs. Sporadically, the virus jumps the species barrier from these domestic animals to humans causing a world-wide pandemic that can infect 30-50% of the population in a single winter season. This is what may happen with the pandemic swine-origin H1N1 virus because few people have protective antibodies against it.
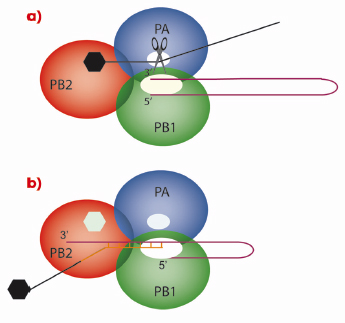
As with all genes, those of influenza virus need to be transcribed into messenger RNA (mRNA) that is subsequently translated into proteins by ribosomes. However, viruses have no metabolism of their own and must use the ribosomes of the cells they have infected. Therefore, the viral mRNA molecules must resemble cellular mRNAs otherwise the ribosomes will not recognise them. One of the characteristics of all cellular mRNAs is that they start with a molecular tag, a “cap”, that is added to the molecule by cellular RNA capping enzymes during the process of transcription of DNA genes into RNAs. Because the genes of influenza virus are made of RNA that cannot be read by the cellular RNA polymerase, the virus encodes its own polymerase that can copy RNA from a RNA template. However, the resulting RNA molecules are uncapped and the viral enzyme cannot use the cellular functions to make a cap structure. This problem is encountered by all RNA viruses and each RNA virus family has found its own solution to modify its mRNA such that it is recognised by host cell ribosomes. Influenza employs perhaps the most devious method of all viruses: cap-snatching! The virus replicates in the nucleus of the infected cell; the place where cellular mRNAs are produced. Its RNA polymerase is a trimer (Figure 117) in which the PB1 subunit carries the actual RNA polymerase function. To make capped viral mRNA, the PB2 subunit binds the cap structure of cellular mRNAs and then an endonuclease activity, revealed in this work to be located in the PA subunit, cleaves the cellular mRNA producing a 10-13 nucleotide, capped RNA primer. This primer is then transferred to the polymerase active site on the PB1 subunit and is extended using the viral genes as templates, thus making capped viral mRNA.
 |
|
Fig. 117: Cap-snatching mechanism of the influenza virus polymerase. In a) the PB2 subunit of the trimeric viral polymerase binds to the cap of a cellular mRNA molecule. The endonuclease activity of PA then cleaves the cellular mRNA which is used in b) by the PB1 subunit as a primer for viral mRNA synthesis. |
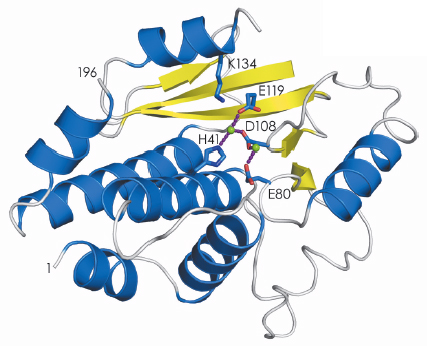
The structure of the active site that binds the cap structure was determined last year [1]. Using data collected at beamlines ID23-1 and ID14-4, we have now determined the structure of the endonuclease domain that belongs to the PD-(D/E)XK nuclease superfamily, revealing an active site containing two metal ions (Figure 118). The biochemical work that led up to the crystallisation of the two active domains also showed that they fold independently and have the same activity as the domains in the intact polymerase trimer. This has opened up the possibility of performing high-throughput screening of small molecules to find inhibitors of cap-binding or endonuclease activities. Before this work such experiments were impossible because complicated and unstable viral polymerase preparations had to be used. Once inhibitors have been identified, co-crystallisation with the individual domains will reveal their binding mode and guide further modifications to produce molecules that bind with higher affinity and specificity. This should contribute to the generation of a new class of anti-influenza drugs that, although arriving too late for the present H1N1 pandemic, will be ready for the next (inevitable) one.
 |
|
Fig. 118: Secondary structure representation of the endonuclease domain of the influenza virus polymerase; ß-sheets in yellow and |
Reference
[1] D. Guilligay, F. Tarendeau, P. Resa-Infante, R. Coloma, T. Crepin, P. Sehr, J. Lewis, R.W.H. Ruigrok, J. Ortin, D.J. Hart and S. Cusack, Nature Struct. Mol. Biol. 15, 500-506 (2008).
Principal publication and authors
A. Dias (a), D. Bouvier (a), T. Crepin (a), A.A. McCarthy (a,b), D.J. Hart (a,b), F. Baudin (a), S. Cusack (a, b) and R.W.H. Ruigrok (a), Nature 458, 914-918 (2009).
(a) Unit of Virus Host Cell Interactions, UMI 3265 UJF-CNRS-EMBL, Grenoble (France)
(b) EMBL Grenoble Outstation, Grenoble (France)



