- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2010
- Structural biology
- Insight into the mechanism of spider silk assembly
Insight into the mechanism of spider silk assembly
Spider silk is known for its remarkable mechanical properties, combining strength, toughness, and extensibility. This high-performance biopolymer is constructed of specific proteins, spidroins, with a repetitive sequence flanked by two different conserved non-repetitive domains. Spiders have evolved an elegant silk production system in which various specialised glands are optimised to fulfil different needs. Some spiders can spin up to seven different types of silk that are used for different purposes, such as protection of the eggs in the egg-case or construction of a capture-spiral. Spidroins are stored as a highly concentrated liquid-crystalline solution (‘dope’) that, upon silk formation, converts into β-sheet rich structures in which inter- and intramolecular interactions between repeat regions convey strength (alanine-blocks) and elasticity (glycine-rich motifs). Understanding the mechanisms underlying spider silk production will facilitate development of silk-based biomaterials, with interesting possibilities for industrial and scientific applications.
 |
|
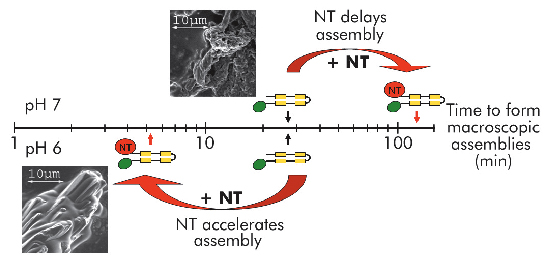
Fig. 94: Self-assembly of mini-spidroins with or without NT at pH 7 and 6 (above and below timescale, respectively). Arrows indicate when macroscopic structures were first detected. Scanning electron micrographs represent early structures of minature spidroins without NT at pH7 (above axis) and of minature spidroins with NT at pH 6 (below axis). |
We recently published work providing insight into how spider silk assembly is controlled and how premature spidroin aggregation prior to silk assembly is avoided. Conversion of spidroins from a highly concentrated protein dope to spider silk is believed to be triggered by gradually changing environmental conditions along the length of the spinning apparatus, including an alteration in ion composition, a lowering of the pH from neutral to below 6.3, retraction of water, and increased shear forces [1]. Recombinant miniature spidroins consisting of a few repetitive spidroin segments capped by the carboxy-terminal domain of major ampullate spidroin (MaSp) 1 from the nursery web spider Euprosthenops australis form metre-long fibres irrespective of pH. We discovered that the amino-terminal domain of MaSp1 (NT) serves as a relay within the physiologically relevant pH range; at neutral pH, NT can be stored at very high concentrations (> 210 mg/ml) without aggregation, but, below pH 6.4, NT forms larger protein assemblies with a hydrodynamic size of ~700 nm. NT assembly is reversible and can be blocked by a pH increase to above 6.4, and/or by introducing 300 mM NaCl. When NT is combined with other parts from the spidroin to generate minispidroins, immediate and irreversible self-assembly occurs at pH values around 6.3, whilst above pH 7 assembly is delayed (Figure 94). This inhibitory effect of NT is even more prominent at high (pH 8) and low (pH 3) pH levels where the minispidroins require days to assemble.
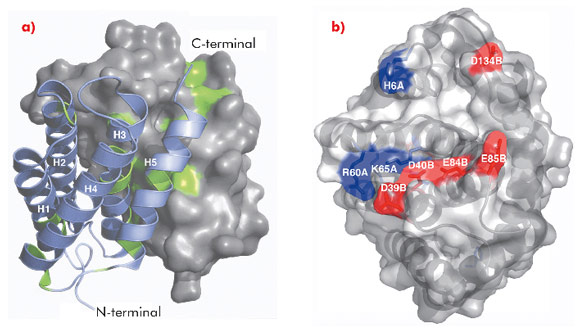
The 1.7 Å X-ray structure of NT, determined using data collected at ID14-2, ID14-4, ID23-1 and ID23-2, revealed a symmetric homodimer of dipolar, antiparallel five-helix bundle subunits without any known structural homologues (Figure 95). The dimer interface is largely hydrophobic, and the interface residues are highly conserved throughout known spidroins. Homology modelling suggests that both the dimerisation and charge distribution observed in NT is conserved for spider silks with widely different mechanical properties and repeat unit sequences. We studied the effects of pH and salt concentration on the assembly process over the entire pH range from pH 3-11. Our results showed that specific local electrostatic interactions, that are available only around pH 6.3, are crucial for functional (wt-like) assembly of minispidroins. The crystal structure of NT revealed numerous conserved acidic residues at or near the dimer surface that could mediate electrostatic assembly interactions. We identified three conserved acidic residues that might play a role in pH-sensitive assembly and probed their role through site directed mutagenesis. We discovered that none of these residues alone are required for determining the pH of the transition from soluble to assembled material, but that two of the residues (D40 and E84) are required for wt-like assembly. Our results present a first insight into the mechanism of spider silk assembly at the atomic level. The pH sensitive relay-like behaviour of NT regulates this process by inhibiting precocious aggregation during storage, and accelerating self-assembly as the pH is lowered along the spider’s silk extrusion duct.
Principal publication and authors
G. Askarieh (a,b), M. Hedhammar (c), K. Nordling (c), A. Saenz (d), C. Casals (d), A. Rising (c), J. Johansson (c) and S.D. Knight (a), Nature 465, 236-238 (2010).
(a) Department of Molecular Biology, SLU, the Biomedical Centre, Uppsala (Sweden)
(b) Department of Chemistry, Oslo University (Norway)
(c) Department of Anatomy, Physiology and Biochemistry, SLU, the Biomedical Centre, Uppsala (Sweden)
(d) Department of Biochemistry and Molecular Biology I & CIBER Enfermedades Respiratorias, Complutense University of Madrid (Spain)
References
[1] A. Rising , M. Widhe, J. Johansson and M. Hedhammar, Cell Mol Life Sci. (2010); DOI:10.1007/s00018-010-0462-z.