- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2008
- Materials science
- Metal-peptide frameworks
Metal-peptide frameworks
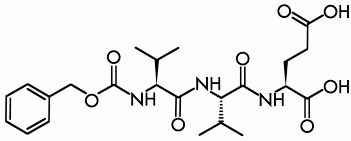
Like zeolites, the class of materials known as metal organic frameworks (MOFs) are both porous and crystalline. However, while a zeolite framework is purely inorganic and is restricted to tetrahedral building units, the MOF coordination polymers are inorganic/organic hybrids and the possibilities for varying the nature of the metal centres and of the organic linkers to induce specific characteristics in the resulting material are innumerable. Interestingly, the possibility of using peptides as connectors has not been reported, although they offer a rich structural diversity and are intrinsically chiral. To explore their potential as linkers in the fabrication of chiral metal peptide frameworks (MPFs), a series of syntheses using Cu2+ and Ca2+ as metal centres and oligovalines as linkers was performed. The combination of Cu2+ and the Z-(L-Val)2-L-Glu(OH)-OH oligopeptide (Figure 41) produced the polycrystalline material “MPF-9”.
 |
|
Fig. 41: The Z-(L-Val)2-L-Glu(OH)-OH oligopeptide. |
Powder diffraction methods had to be used for structure solution because there were no single crystals of the product. To this end, high-resolution powder diffraction data were collected using the diffractometer on BM01B (Swiss-Norwegian Beamline). The pattern could be indexed with a monoclinic unit cell (a = 19.2610 Å, b = 4.8885 Å, c = 30.5096 Å, ß = 92.524°) and the systematic absences were consistent with the space groups C2, C2/m or Cm. As a non-centrosymmetric structure was expected, C2 was assumed for structure analysis.
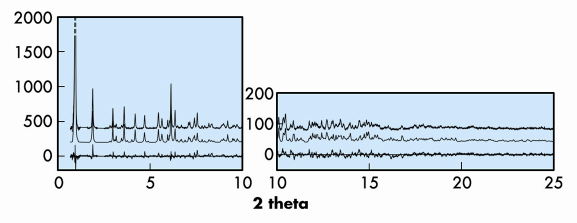
The data proved to be of only modest resolution in reciprocal space (dmin of 1.4 Å), but considerable chemical information about the peptide structure and the Cu coordination geometry was known, so a direct-space approach to structure solution was attempted. For this purpose, the direct-space global-optimisation algorithms implemented in the computer program FOX [1] were used. A simplified structural model with limited conformational degrees of freedom was used as input, and the model with the best fit to the data was then taken as a starting point for structure completion and Rietveld refinement. For the latter, the complete pattern was used, and bond distance and angle restraints were included to keep the model chemically sensible during the course of the refinement. The profile fit for the final model (RF = 0.103, Rwp = 0.266, Rexp = 0.217) is shown in Figure 42.
 |
|
Fig. 42: Observed (top), calculated (middle) and difference (bottom) profiles for the Rietveld refinement of MPF-9. The first peak has been cut at half height to show more detail. |
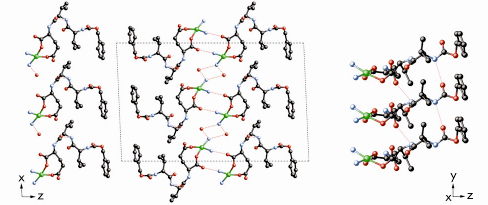
The Cu2+ ions in the structure are each coordinated to two oxygen atoms of the peptide carboxylate groups and to two ammonia molecules in a distorted square planar geometry (Figure 43). One of the coordinated NH3 molecules forms a hydrogen bond with the L-glutamic acid residue of the neighbouring complex along the [001] direction, while the other forms hydrogen bonds with two water molecules, which form a helical hydrogen bonding network along the [010] direction. The peptide ligands themselves also form hydrogen bonds to one another along the [010] direction to create a ß-sheet-like structure. This extended hydrogen-bonding network accounts for the insolubility of MPF-9– in both water and organic solvents, and the concomitant difficulty in growing single crystals.
 |
|
Fig. 43: Projections of the structure of MPF-9 along [010] (left) and along [110] (right) showing the H-bonding network. |
The structure is, within the limits of the method, very well defined. It is also consistent with the results of the elemental analysis, TGA, EPR and magnetic measurements. It can be described in terms of two 2-dimensional substructures. The first of these consists of a double layer of copper ions in the ab plane, in which the two layers are diagonally displaced with respect to one another, and the second is that of the peptide with the long axis of the molecule oriented roughly along [001]. In addition to the hydrogen bonding network along the [010] direction, the phenyl groups of neighbouring peptides along [001] form columns along [010] and are oriented to allow ![]() -
-![]() interactions.
interactions.
Although MPF-9 does not have an extended 3-dimensional Cu-peptide network, magnetic interactions were found to occur between magnetic centres; the structure does have some porosity and the peptide has been shown to be a viable ligand meriting further investigation.
Principal publication and authors
A. Mantion (a), L. Massüger (b), P. Rabu (c), C. Palivan (a), L.B. McCusker (b), A. Taubert (a,d), J. Am. Chem. Soc., 130, 2517 (2008).
(a) Department of Chemistry, University of Basel (Switzerland)
(b) Laboratory of Crystallography, ETH Zurich (Switzerland)
(c) IPCMS, UMR 7504 CNRS - Université Louis Pasteur (Strasbourg)
(d) Institute of Chemistry, University of Potsdam (Germany)
References
[1] V.R. Favre-Nicolin, R. Cerny, J. Appl. Crystallogr. 35, 734-743 (2002).



