- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2008
- Structural biology
- Structural basis of Dcp2 recognition and activation by Dcp1
Structural basis of Dcp2 recognition and activation by Dcp1
RNA degradation plays an important role in the control of gene expression and in the elimination of aberrant mRNAs. Decapping is a critical step since it promotes the bulk of mRNA turnover and functions in specialised pathways such as nonsense-mediated decay (NMD), adenine/uridine-rich element (ARE)-mediated decay, and the turnover of some mRNAs promoted by miRNAs [1].
Removal of the 5’ cap structure is catalysed by the decapping complex consisting of at least two subunits: Dcp1p and Dcp2p. Dcp1p is a small protein containing an EVH1 domain, which is generally a protein-protein interaction module. The catalytic subunit, Dcp2p, recognises capped mRNA substrates and cleaves the pyrophosphate bond of the m7GpppN cap, releasing m7GDP as product. Dcp2p belongs to the nudix hydrolyase family, which contain the nudix signature nudix motif. The crystal structure of Dcp2p from S. pombe reveals that a conserved N-terminal region forms a two-lobed structure with an α-helical domain preceding the nudix domain [2]. That Dcp2p prefers longer RNA substrates suggests the presence of spatially separated RNA-binding and catalytic sites on the nudix domain, but the nature and site of the RNA binding domain of Dcp2p has not yet been determined. Additionally, despite the conservation of Dcp1p and its requirement for decapping, how Dcp1p interacts with and activates Dcp2p remains elusive.
 |
|
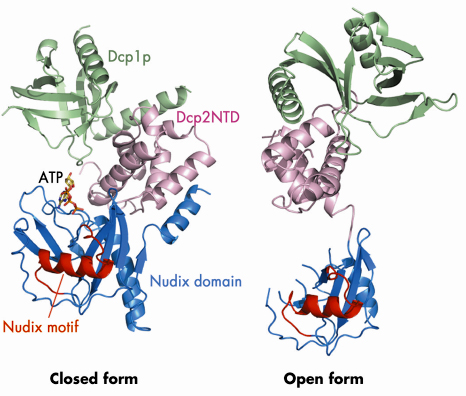
Fig. 65: Ribbon diagrams of the Dcp1p-Dcp2n complex in the closed and open conformations. Dcp1p is shown in green, the N-terminal domain (NTD) of Dcp2n in pink, the nudix domain of Dcp2n in blue and its nudix motif in red. In the closed conformation, the ATP molecule we observe in the active site is shown in a stick representation. |
We determined the crystal structure of Dcp1p in complex with a truncated S. pombe Dcp2p (Dcp2n) using data collected on beamline ID29. The structure reveals both open and closed forms of the Dcp1p/Dcp2n complex (Figure 65). In the closed complex, a compact structure is formed with both Dcp1p and the N-terminal ![]() -helical domain of Dcp2n being in close proximity to the nudix domain of the latter. In the open form, the
-helical domain of Dcp2n being in close proximity to the nudix domain of the latter. In the open form, the ![]() -helical and nudix domains of Dcp2n do not interact and are separated by a flexible linker, making the whole molecule resemble a dumb-bell. Small-angle X-ray scattering (SAXS) analysis confirmed that the two forms of the Dcp1p/Dcp2n complex also exist in solution and that formation of the closed complex is influenced by ligand binding. Interestingly, we observed that ATP is bound to the catalytic centre (Figure 65), and that ATP, di-nucleotide (pAA) and the cap analogue m7GpppA enhance the formation of the closed complex and inhibit decapping. Mutational analysis in the flexible linker region indicated that the closed complex we observe is, or closely resembles, the catalytically more active form, suggesting that a conformational change between open and closed complexes might control decapping. We have also identified a region involved in RNA binding which is located some distance from the active site (Figure 66).
-helical and nudix domains of Dcp2n do not interact and are separated by a flexible linker, making the whole molecule resemble a dumb-bell. Small-angle X-ray scattering (SAXS) analysis confirmed that the two forms of the Dcp1p/Dcp2n complex also exist in solution and that formation of the closed complex is influenced by ligand binding. Interestingly, we observed that ATP is bound to the catalytic centre (Figure 65), and that ATP, di-nucleotide (pAA) and the cap analogue m7GpppA enhance the formation of the closed complex and inhibit decapping. Mutational analysis in the flexible linker region indicated that the closed complex we observe is, or closely resembles, the catalytically more active form, suggesting that a conformational change between open and closed complexes might control decapping. We have also identified a region involved in RNA binding which is located some distance from the active site (Figure 66).
 |
|
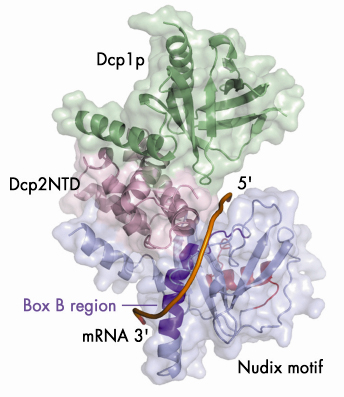
Fig. 66: A model illustrating the putative RNA binding channel in the closed form of the Dcp1p-Dcp2n complex. A 12-mer poly(A) mRNA is shown in a tube representation and the Box B region of Dcp2n is coloured in dark purple. |
Our structural analyses coupled with biochemical data suggest a possible mechanism by which Dcp2p recognises its substrate. The substrate is bound in a channel on the surface of Dcp2p with the cap structure in the active site (Figure 66). The main body of the RNA then wraps across the nudix domain and follows the channel along the Box B helix. This model provides a possible structural explanation for the preference of decapping enzymes for longer RNA substrates: RNA would need to be at least 12 residues long to bind in both the active site and the Box-B region on Dcp2p.
Our analyses suggest that Dcp1p stimulates the activity of Dcp2p by stabilising the closed complex, which is promoted by the presence of substrate in the active site. Finally our structure also reveals that the Dcp1/Dcp2 interface is not fully conserved, explaining why the Dcp1-Dcp2 interaction in higher eukaryotes requires an additional factor.
Principal publication and authors
M. She (a), C.J. Decker (b), D.I. Svergun (c), A. Round (c), N. Chen (a), D. Muhlrad (b), R. Parker (b), H. Song (a), Mol. Cell 29, 337 (2008).
(a) Laboratory of Macromolecular Structure, Institute of Molecular and Cell Biology (Singapore)
(b) Department of Molecular and Cellular Biology and Howard Hughes Medical Institute, University of Arizona (USA)
(c) Hamburg Outstation, EMBL (Germany)
References
[1] J. Coller, and R. Parker, Annu Rev Biochem 73, 861 (2004).
[2] M. She, C.J. Decker, N. Chen, S. Tumati, R. Parker, and H. Song, (2006). Nat Struct Mol Biol 13, 63 (2006).



