- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2003
- Macromolecular Crystallography
- Cytochrome P450 2C9 with Bound S-warfarin
Cytochrome P450 2C9 with Bound S-warfarin
Cytochromes P450 are haem-containing enzymes found in many bacteria, fungi, plants and animals. P450s catalyse the biotransformation of a wide range of structurally-diverse endogenous and exogenous substrates. A number of human cytochromes P450 are of great interest to the pharmaceutical industry, due to their role in phase I metabolism of drugs. During phase I the reaction most commonly catalysed by P450s is the hydroxylation reaction
RH + O2 + 2H+ + 2e- ![]() ROH + H2O
ROH + H2O
The electrons required are donated by NADPH-cytochrome P450 reductase and cytochrome b5. In phase II metabolism this group is then conjugated, thus increasing the solubility of the compound and facilitating excretion. By understanding the way in which P450s recognise and metabolise compounds, it is hoped that modifying these interactions may alter the pharmaceutical profile of compounds.
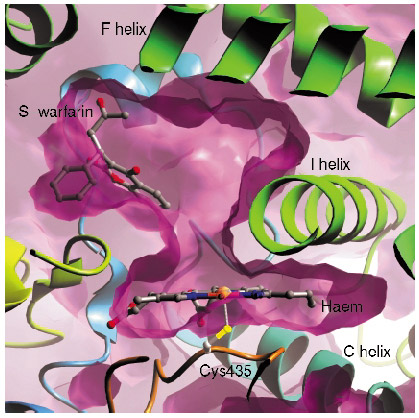
In contrast with many of the bacterial P450s whose structures have been solved, the mammalian drug-metabolising P450 enzymes are membrane-associated proteins, and this hampered production of protein suitable for structural studies. We have generated an active, truncated 2C9 protein that crystallises and diffracts to a resolution of 2.6 Å. We collected data from P450 2C9 crystals on ESRF beamline ID14-1 to 2.6 Å resolution and solved the structure by molecular replacement. The haem group forms the bottom of a large active site, with different regions of the polypeptide chain forming the active site. We then went on to solve the structure of 2C9 in complex with the anticoagulant substrate, S-warfarin, using data collected on beamline ID14-2. Contrary to expectations, the S-warfarin binding site is some 10 Å away from the haem (Figure 27). The location of the binding site suggests an allosteric mechanism, and leaves sufficient room directly above the haem for an additional compound molecule to bind, giving a structural basis for drug-drug interactions.
 |
|
Fig. 27: The binding site of S-warfarin within the active site of 2C9 leaves the haem, shown edge on in the figure, accessible to other compounds. |
Principal Publication and Authors
P.A. Williams (a), J. Cosme (a), A. Ward (a,b), H.C. Angove (a), D. Matak Vinkovic (a), H. Jhoti (a), Nature 424, 464-468 (2003).
(a) Astex Technology, Cambridge (UK)
(b) Current address: AstraZeneca, R&D Charnwood, Loughborough (UK)



