- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2003
- Macromolecular Crystallography
- Further Steps Towards Beamline Automation
Further Steps Towards Beamline Automation
Reviewed by J. McCarthy and W. Shepard, for the Joint Structural Biology Group (JSBG) of the ESRF and the EMBL Grenoble Outstation.
Macromolecular crystallography (MX) has proven to be the most effective method of determining the structures of biological macromolecules. Each year thousands of data collections that form the basis for such structure determinations are carried out at the ESRF. To cope with this high demand, it is essential to maximise beamline efficiency and to simplify beamline control procedures. We are therefore automating much of the crystallographic experiment which may be divided into two parts: firstly beamline and crystal alignment, and secondly data collection and processing.
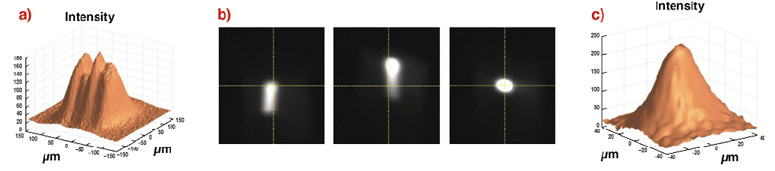
Beamline setup is extremely important since many crystals of biological macromolecules do not grow much larger than tens of micrometres in size. Often the X-rays need to be micro-focussed into a small uniform spot. An elegant and automated procedure based on wavefront analysis has been developed at the ESRF using a position sensitive X-ray camera. This method, called Automatic Beamline Alignment (ABA), allows the correct orientation of the X-ray optics to be deduced from the measurement of only a few parameters. Figure 20 shows the unfocussed and ABA micro-focussed beam on ID29.
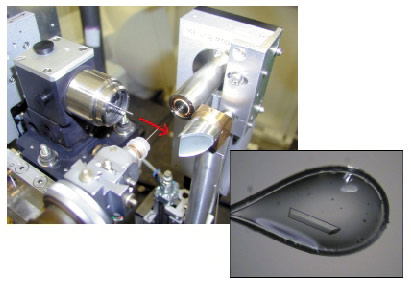
Aligning micrometre-sized crystals in a micro-focussed X-ray beam has been facilitated by a special microscope that allows visualisation along the axis of the X-ray beam, Figure 21. The position of the X-rays can be directly observed and the sample aligned appropriately. This device is simple, modular and can be adapted to virtually any experimental setup. A prototype has been installed on ID29 and production models will be installed on the ID14 beamlines in early 2004. New software for centring the crystal sample in the beam has been used on ID14-3 since May 2003. This software allows the crystal to be centred using a 3-click procedure via a graphical interface. Software for fully automatic centring of crystals in the beam is currently under development.
 |
|
Fig. 21: On-axis sample visualisation (top left) allows much-improved viewing of small crystals. The red arrow indicates the X-ray beam path. |
A sample changer robot, which can hold up to 50 frozen samples, was installed on ID14-3 in December 2003. Additionally, the DNA software that allows for automatic crystal characterisation is now available on all MX beamlines. Once fully implemented, the combination of sample changer robot, automatic sample centring protocols and DNA software will mean virtually 'hands-free' beamlines that will allow crystal screening and data collection processes to be carried out rapidly and efficiently.
Techniques exploiting anomalous diffraction (MAD, SAD) are pre-eminent for the de novo determination of macromolecular structures. The correct choice of wavelength for data collection based on a scan of the absorption edge of a heavy atom in the crystal is a crucial first step. These calculations have been automated on ID14-4 and ID29. This 'one-click' procedure known as AEscans, carries out all necessary configuration of X-ray source and beam; optimises X-ray beam attenuation; performs a XANES scan; analyses the scan to deduce the correct wavelengths then passes this to the data collection interface. MAD and SAD experiments are thus simple and straightforward to carry out at the ESRF.
The tools described above should, in the coming year, help to provide an efficient and effective environment for high-throughput molecular crystallography that should be of great benefit for the user community.