- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2002
- Macromolecular Crystallography
- Structure of the Electron-transfer Complex of Cytochrome c and Cytochrome bc1 Complex from Yeast
Structure of the Electron-transfer Complex of Cytochrome c and Cytochrome bc1 Complex from Yeast
Electron transfer processes are of great importance in many metabolic pathways of living organisms. They are essential for respiration, a process in which energy gained from oxidation of nutrients is converted to ATP. Energy conversion is achieved by coupling the transfer of electrons along a chain of proteins to the translocation of protons across a lipid membrane. The resulting electrochemical proton gradient can then be used for ATP synthesis. In the respiratory chain of eukaryotic cells electron transfer involves four multisubunit complexes that are embedded in the inner mitochondrial membrane. Electrons are transferred between two of these, namely the cytochrome bc1 complex and the cytochrome c oxidase, via reversible docking of the small soluble protein cytochrome c (CYC).
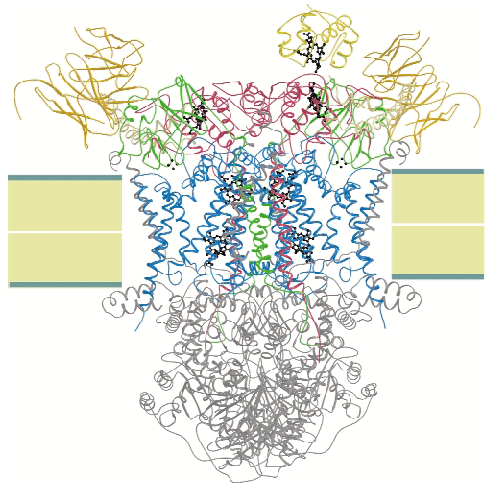
We crystallised the complex of cytochrome c and cytochrome bc1 complex (QCR) from the yeast Saccharomyces cerevisiae with the help of an antibody Fv fragment. The molecular structure of this entity was determined at 2.97 Å resolution by molecular replacement using the previously determined high resolution structure of QCR [1] as search model (Figure 3). This is the first structure of a complex of redox partners from the mitochondrial respiratory chain.
 |
|
|
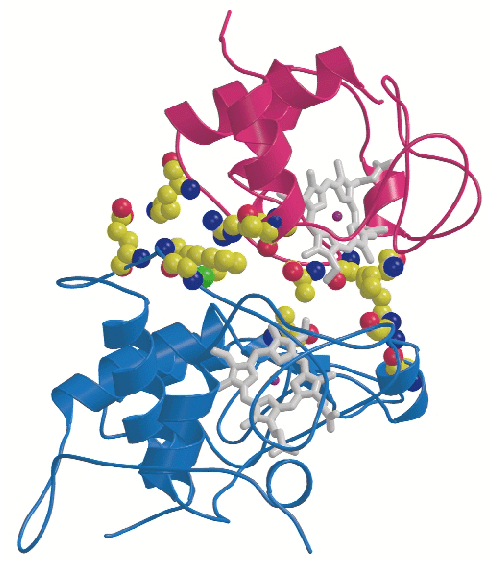
CYC binds to the cytochrome c1 subunit of the cytochrome bc1 complex. The resulting complex between the two is stabilised by hydrophobic interactions surrounding the heme crevices thereby creating a small, compact contact site (Figure 4). A central cation- interaction is an important and conserved feature of CYC binding [2] and peripheral patches with highly-conserved complementary charges further stabilise the enzyme-substrate complex using long-range electrostatic forces that may also serve to ensure the orientation of CYC. The close spatial arrangement of the hemes suggests a direct and rapid heme-to-heme electron transfer at a rate of ~8 x 106 sec1. CYC reduction by QCR is highly dependent on ionic strength with an optimum activity at physiological ionic strength [2]. As the crystals of the complex were obtained at physiological ionic strength, and as the size and characteristics of the contact site are optimal for the formation of a transient complex allowing fast electron transfer, there is strong indication that the structure represents the true native electron transfer complex. Remarkably, CYC binds only to one of the two possible binding sites of the homodimeric complex and binding is correlated with the presence of ubiquinone at the site of quinone reduction. This suggests a regulated binding of ubiquinone and CYC which act as electron acceptors for the two-electron oxidation of the substrate ubiquinol.
interaction is an important and conserved feature of CYC binding [2] and peripheral patches with highly-conserved complementary charges further stabilise the enzyme-substrate complex using long-range electrostatic forces that may also serve to ensure the orientation of CYC. The close spatial arrangement of the hemes suggests a direct and rapid heme-to-heme electron transfer at a rate of ~8 x 106 sec1. CYC reduction by QCR is highly dependent on ionic strength with an optimum activity at physiological ionic strength [2]. As the crystals of the complex were obtained at physiological ionic strength, and as the size and characteristics of the contact site are optimal for the formation of a transient complex allowing fast electron transfer, there is strong indication that the structure represents the true native electron transfer complex. Remarkably, CYC binds only to one of the two possible binding sites of the homodimeric complex and binding is correlated with the presence of ubiquinone at the site of quinone reduction. This suggests a regulated binding of ubiquinone and CYC which act as electron acceptors for the two-electron oxidation of the substrate ubiquinol.
 |
|
Fig. 4: Close-up view showing binding interactions between cytochrome c (purple) and the cytochrome c1subunit (blue) of the bc1 complex. Side chains of amino acid residues involved in stabilising interactions are shown in a space-filling representation. |
References
[1] C. Hunte, J. Köpke, C. Lange, T. Roßmanith and H. Michel, Structure 8, 669-684 (2000).
[2] C. Hunte, S. Solmaz and C. Lange, Biochim. Biophys. Acta 1555, 21-28 (2002).
Principal Publication and Authors
C. Lange and C. Hunte, Proc. Natl. Acad. Sci. USA 99, 2800-2805 (2002).
Dept. Molecular Membrane Biology, MPI, Frankfurt (Germany)



