- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2001
- Life Sciences
- Structure of an Enveloped Virus: the Semliki Forest Virus
Structure of an Enveloped Virus: the Semliki Forest Virus
Amongst the wide variety of viruses, some have rather simple spherical structures. Examples are the rhinovirus, which causes the common cold, and the poliovirus, the agent causing poliomyelitis. They consist of a single protein shell surrounding a nucleic acid molecule that carries the viral genome information. These are called non-enveloped viruses and their mode of infection involves an attachment step whereby their cellular receptors link to the host cell, followed directly by the injection of their viral genome into the cell and, subsequently, by multiplication of the virus.
In contrast, enveloped viruses, which are multi-layered structures composed of a series of concentric protein shells and one lipid envelope, have a specific step before infection, namely the fusion (or merging) of the viral and host cell membranes. In the case of the Semliki Forest Virus (SFV), an alphavirus, which can provoke encephalitis and which is transmitted to humans by mosquitoes, the fusion of the host cell and the viral membranes is induced by a single protein called E1. The E1 protein is embedded in the lipid envelope of the virus and, together with a second viral protein named E2, forms spikes on the outer surface of the viral particle. During infection, the E2 protein is believed to bind to a host cell receptor and this leads to the entry of the viral particle into the host cell. When SFV is exposed to the acidic pH of the endosome within the cell, the conformation of the E1 protein changes drastically. This event leads to membrane fusion and allows the infectious cycle to proceed.
X-ray crystallographic data collected on beamlines ID2 and ID14 have allowed the determination of the 3D structure of the E1 protein at neutral pH. A combination of this data with Electron Microscopy [1], allowed the reconstruction of the fusion shell of the entire virus particle (Figure 5). These results have highlighted the crucial scaffolding role of the E1 protein to form a closed virus particle and thus, to direct the exit of newly formed viral particles from the host cell.
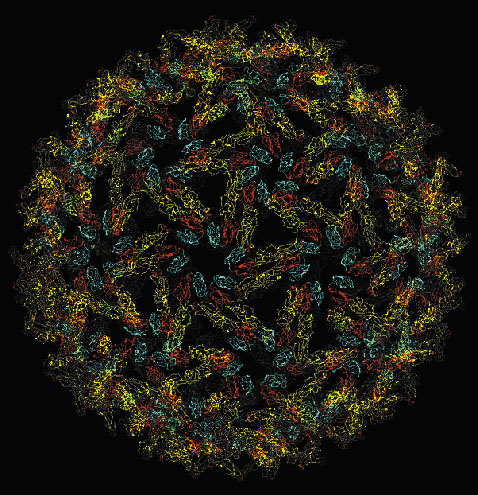 |
Fig. 5: The T = 4 icosahedral protein layer formed by E1 on the Semliki Forest Virus surface. |
SFV is currently used to selectively express some proteins in susceptible host cells and could therefore be of use in some gene therapy protocols. Virologists are also using the Semliki Forest Virus particle as a scaffold to present viral envelope proteins to the immune system. With the help of the 3D structure of SFV, one could therefore envision preparing chimerical viral particles for vaccination. Finally and quite unexpectedly, it turned out that the structure of the SFV E1 envelope protein is very similar to the structure of the envelope protein from Tick Borne Encephalitis, a member of the flaviviridae family of viruses [2]. This family includes important human pathogens like Yellow-Fever Virus or Dengue Virus, with no vaccine yet being available for the latter. Knowledge of the structure of Semliki Forest Virus suggests that alphaviruses and flaviviruses use a common mechanism of infection and may help in providing some possible strategies to prevent it.
References
[1] E.J. Mancini, M. Clarke, B.E. Gowen, T. Rutten and S.D. Fuller, Mol. Cell 5, 255-266 (2000).
[2] F.A. Rey, F.X. Heinz, C. Mandl, C. Kunz and S. C. Harrison, Nature 375, 291-298 (1995).
Principal Publications and Authors
J. Lescar (a), A. Roussel (b), M.W. Wien (b), J. Navaza (b), S.D. Fuller (c), G. Wengler (d) and F.A. Rey (b), Cell., 105, 137-148 (2001); G. Wengler (d) and F.A. Rey (b), Virology 257, 472-482 (1999).
(a) ESRF
(b) Laboratoire de Génétique des Virus, C.N.R.S.-UPR9053 (France)
(c) University of Oxford (UK)
(d) Institut für Virologie, Giessen (Germany)



