- Home
- News
- General News
- Stress-adapted proteins...
Stress-adapted proteins resist in adverse conditions
15-05-2007
PRESS RELEASE- Around 30% of human tumours arise due to mutations that hamper the cells ability to control and regulate key pathways, which channel growth and other signals from the cell surface to the nucleus. Scientists around the world are studying the mechanisms that underpin these pathways and the proteins components from which they are constructed. A team from Cancer Research UK has discovered a new family of proteins that regulate stress pathways in yeast, and they have determined their 3D structure by using synchrotron light at the ESRF.
Share
Stress response pathways are triggered by a diverse range of stimuli that could be harmful to the cell, including potentially dangerous free radicals, UV radiation and heat shock. The pathways work through the step-by-step activation of specialised proteins, which transmit stress signals detected at the cell surface through to the nucleus, where the cellular response can be coordinated. One of the key pathways involved in alerting the cell to the presence of stress signals in its environment is the Mitogen-Activated Protein (MAP) kinase pathway.
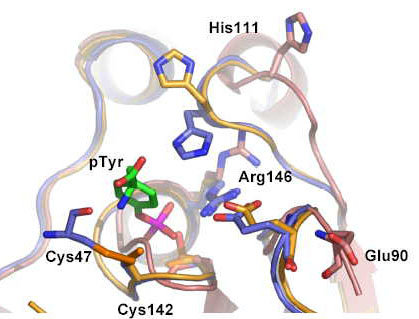 |
|
The active site of Sdp1 showing both the high and low activity states. The key disulphide between Cysteine 47 and Cysteine 142 which mediates the oxidative activation mechanism is shown in gold. |
In recent years, scientists have become aware of the important role played by redox-mediated reactions (reactions that involve changes in oxidation state) in influencing and regulating cellular pathways. It has also become clear that if the cell loses control of pathways such as the MAP kinase pathway, it can have disastrous consequences - turning normal healthy cells into cancer cells.
The team from Cancer Research UK, headed by Prof Neil McDonald, has identified a protein phosphatase called Sdp1, which acts on a MAP kinase pathway activated by stress in yeast cells. The team focused on this protein because it is produced when cells come under attack by oxidative free radicals. “Sdp1 has to be able to operate in an environment rich in free radicals that would normally inactivate this type of protein”, explains Gavin Fox, first author of the paper. “We set out to answer how this enzyme has evolved the ability to operate and remain active under these conditions”.
Scientists carried out experiments at the ESRF to determine the 3D structure of the protein in different oxidation states and used this information to unravel the detailed molecular mechanism of Sdp1. “We needed a combination of techniques: protein crystallography, enzyme kinetics, mutational studies and sophisticated mass spectrometry, before we could put together a detailed picture of what was going on”, explains Fox.
The researchers have discovered that Sdp1 operates through a novel “oxidative-activation” mechanism that uses the formation of a disulphide-bond as a switch to boost its activity.
The research does not end here. The team have now identified a family of similar proteins in yeast and filamentous fungi that are likely to work through the same mechanism. They have named this previously unrecognised family of proteins, the WH phosphatases.
Fox et al., Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatises, Nature (9 May 2007).
For more information, please contact Montserrat Capellas ( email or +33476882663).



