- Home
- News
- Spotlight on Science
- Toxoplasma’s immunogenic...
Toxoplasma’s immunogenic balancing act explained
14-03-2017
The parasite Toxoplasma gondii is a silent success. It infects up to 95% of people in many regions of the world, and most of them never know it due to the parasite’s manipulation of its host’s immune response. Structural analysis of a protein secreted by the parasite has demonstrated exactly how it rewires a crucial signalling network in the human immune response.
When a pathogen invades its host cell it must do its best to avoid the defence mechanisms that have been put in place to repel it. This is an arms race that has been raging between viruses, bacteria and eukaryotic parasites and their targets for millions of years. However, it is a delicate procedure as the invader must also avoid killing its host by being too successful. The eukaryotic parasite Toxoplasma gondii is a master at regulating its host’s defences. Over half the world’s population are infected but show no symptoms. Toxoplasma manages this by keeping the immune response of its host low enough so that it can thrive, but high enough so that its human hosts generally live healthy lives and can incubate parasites. However, for people with a compromised immune system, such as those undergoing chemotherapy, infected with HIV or with organ transplants, toxoplasmosis can be deadly. Additionally, if a mother is infected during pregnancy there can be serious consequences for the developing foetus. Combined with the fact that toxoplasmosis is a close relative of the causative agent of malaria, the study of this parasite is important.
On detection of a parasite, a host cell sets off a chain reaction where a series of molecules activate each other until a protein, called p38α, is activated and moves into the nucleus. Human p38α is a type of protein called a kinase that can transmit a signal and activate targets by placing a phosphoryl group at a specific location on a target protein. In the nucleus, it turns on the proteins that trigger the inflammatory response. Among other things, the purpose of this response is to eliminate the pathogen. In order to survive, toxoplasmosis must modulate the response. Recently, a protein, called GRA24, was discovered that is excreted by toxoplasmosis shortly after invading human cells and was found to co-locate to the nucleus with human p38α [1]. Using a wide variety of techniques available at the ESRF and the EPN campus, the structure of a complex between GRA24 and p38α has now been solved, allowing its mode of action to be determined.
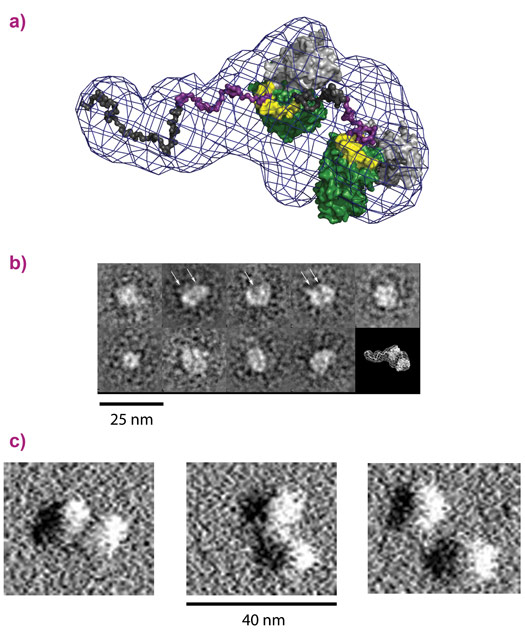
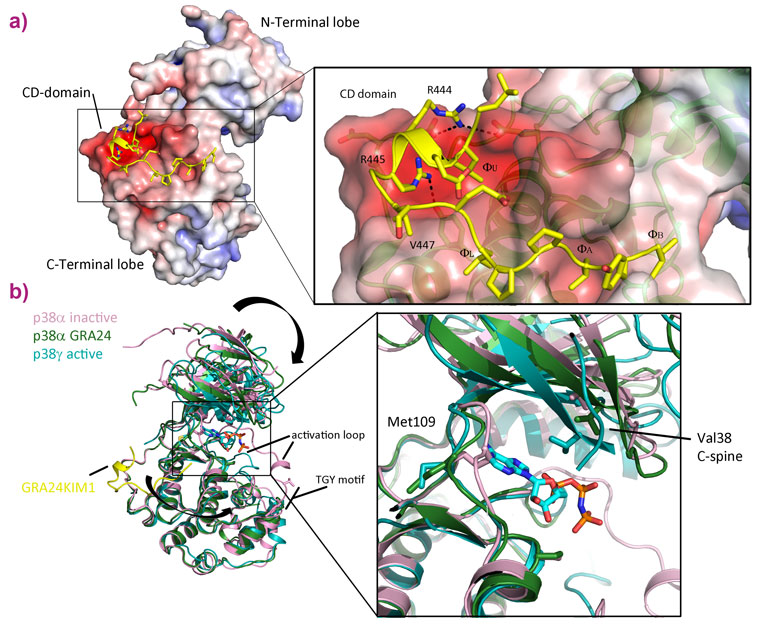
To study the mode of action of the toxoplasmosis protein, we produced the human and parasite proteins together in bacteria. This enabled us to purify large amounts of a complex comprising the two proteins and investigate its properties biochemically and structurally. GRA24 was predicted to belong to a class of proteins called intrinsically disordered proteins (IDPs) that do not fold and are extremely flexible. This property makes crystallography difficult so we turned to several alternative structural techniques available at the ESRF and the EPN campus: SAXS at BM29, AFM at the PSCM surface science lab and electron microscopy at the PSB. These techniques revealed the structure of the complex, confirming that GRA24 is completely disordered and binds two molecules of the human protein p38α that are observed in a large number of conformations (Figure 1). These results shed light on how the complex is formed and acts but gave no information on the molecular details of interaction between the proteins or how activation occurs. We then returned to X-ray crystallography to study the core interaction between the proteins. Crystals of the complex were obtained but varied enormously in their diffraction quality. In order to collect the best data, the ID23-2 microfocussed beam combined with the latest developments in X-ray based centring were essential [2]. The structure revealed that toxoplasmosis has adapted a specific interaction motif, called a KIM, that is normally used in human signalling networks. By combining features from these KIMs, the parasite is able to latch onto p38α and hijack the protein. The binding is also able to activate the kinase by changing its conformation (Figure 2).
By bringing together multiple techniques, the form, function and mechanism of action of this rewiring of the signalling pathway has been unveiled. After invasion, toxoplasmosis secretes GRA24 into the host cell cytoplasm. The high affinity KIM motif is then able to compete with those of the natural signalling network to hijack p38α. Two molecules of p38α are bound and activated forming a highly active form of the kinase ready to be transported into the host nucleus. Once in the nucleus, the complex induces a sustained and constant inflammatory response that prevents eradication of the parasite or host cell death. By using an IDP, toxoplasmosis can adapt very quickly, on an evolutionary timescale, as folded proteins take much longer to adjust and evolve. This research has also generated a new way to assess the efficacy of anti-inflammatory drugs, many of which are designed to block p38α. It is often difficult to evaluate how effective and specific these drugs are as it is hard to produce an active form of p38α in the lab. GRA24 bound to p38α has created a useful tool to do just that. The tight interaction with the parasite protein keeps p38α in its active state, so researchers can now subject it to potential new drugs, and evaluate how well they block p38α’s active site, which the Toxoplasma protein does not interfere with.
To study such a complex system, a multidisciplinary approach was essential. The world-leading beamlines at the ESRF, in combination with the partnerships that have been put in place to support cross-discipline research, were critical in enabling the resolution of this parasite control mechanism.
Principal publication and authors
Structural basis for the subversion of MAP kinase signalling by an intrinsically disordered parasite secreted agonist, E. Pellegrini (a), A. Palencia (b), L. Braun (b), U. Kapp (c), A. Bougdour (b), H. Belrhali (a), M.W. Bowler (a) and M-A. Hakimi (b), Structure 25, 16-26 (2017); doi: 10.1016/j.str.2016.10.011.
(a) European Molecular Biology Laboratory, Grenoble (France)
(b) Institute of Advanced Bioscience, Université Grenoble Alpes, Grenoble (France)
(c) ESRF
References
[1] L. Braun et al., J. Exp. Med. 210, 2071-2086 (2013); doi: 10.1084/jem.20130103.
[2] O. Svensson et al., Acta Cryst D. 71, 1757-1767 (2015); doi: 10.1107/S1399004715011918.