- Home
- News
- Spotlight on Science
- Structural insights...
Structural insights into RNA-mediated transcription regulation in bacteria
07-12-2022
Transcription of RNA from DNA is tightly controlled. Using cryo-electron microscopy (cryo-EM) at beamline CM01, molecular models of RNA polymerase regulated not by other proteins but by the transcript itself were determined. These results shed light on regulatory RNA elements promoting or terminating their own synthesis.
Experiments conducted at cryo-EM beamline CM01 crucially contributed to understanding how regulatory RNA elements are able to modulate RNA polymerase (RNAP) and thus the very enzyme responsible for its own synthesis. Nucleic acids come in two flavours: DNA encodes the genetic blueprint, whereas RNA serves as a labile copy to translate the genetic information into proteins. In addition, RNA has many regulatory and catalytic important roles. RNAP is the enzyme that synthesises RNA from the DNA template. This step is highly regulated and the underlying mechanism is conserved from bacteria to humans. Transcription goes through three phases: i) RNAP binds DNA; ii) RNAP synthesises RNA directed by the DNA template; and iii) RNAP responds to a termination signal, releases the RNA transcript and dissociates from DNA.
Each of these phases is subject to regulation, and a vast number of protein transcription factors modulate RNAP during the process. However, a potentially more ancient and less well-understood mechanism is the regulation of RNAP by the RNA transcript itself, as opposed to regulation by auxiliary protein factors. For example, so-called intrinsic terminators consist of short U-rich RNA sequences that signal RNAP to pause transcription and release the transcript. In bacteria, the RNA transcript additionally contains an RNA hairpin structure that accelerates RNA release. Hairpins are not a universal requirement, and transcription termination by RNAP and U-rich signals occurs without the need for it in eukaryotes, and in the absence of additional protein factors. RNA is even more versatile than that, because rather than terminating its own synthesis, it can also promote transcription and prevent termination. For example, a regulatory RNA called putL suppresses transcriptional pausing by RNAP, prevents transcription termination and stimulates its own synthesis.
Click to enlarge
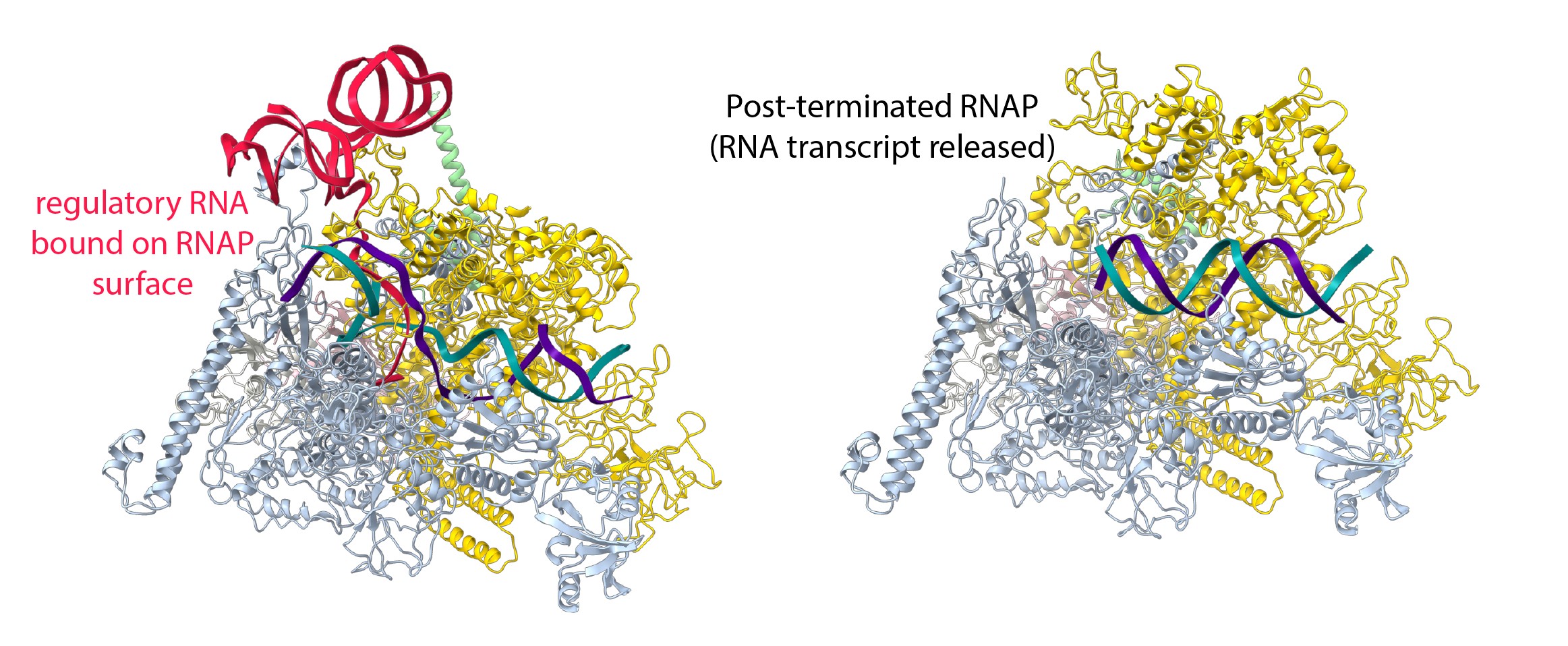
Fig. 1: The structure of RNAP in the presence and absence of the regulatory RNA. Left: Single-particle cryo-EM reconstruction of RNAP trapped at a pause site and transcribing a regulatory RNA, which promotes its own synthesis, provides novel insights into the regulatory versatility of RNA. The regulatory RNA (red) folds into a complex structure, binds the surface of RNA polymerase and favours the active conformation of RNA polymerase. It suppresses pausing and stimulates its own synthesis. Right: In the absence of the regulatory RNA, RNAP could be visualised for the first time after transcript release but before dissociation from the DNA (green and purple).
This work investigated how putL is able to do this without the need for additional protein factors. RNAP was trapped during the synthesis of putL at a U-rich pause signal. Single-particle cryo-EM at beamline CM01 was used to visualise the process at high resolution (Figure 1, left). Structural reconstruction made it possible to build and interpret an atomic model. The regulatory putL RNA binds RNAP in a way that stabilises the active form of the enzyme and disfavours adoption of the paused state. This was confirmed by obtaining structures with an altered putL RNA that lost its functionality. It was observed that RNAP adopts a pause-prone conformation at the U-rich signal. Unexpectedly, it was also found that a subset of RNAP molecules adopted a conformation that appeared to be trapped just before RNA transcript release. To confirm this, another set of cryo-EM reconstructions were determined in the absence of the regulatory RNA, and it was possible to visualise RNAP for the first time before and after RNA transcript release (Figure 1, right).
The set of cryo-EM structures provide a framework to understand how RNA is capable of stimulating its own synthesis. They further highlight intermediate states that occur during intrinsic transcription termination, a universally employed mechanism to release finished RNA products from RNAP.
Principal publication and authors
Structural insights into RNA-mediated transcription regulation in bacteria, S. Dey (a,b,c,d), C. Batisse (a,b,c,d), J. Shukla (a,b,c,d), M.W. Webster (a,b,c,d), M. Takacs (a,b,c,d), C. Saint-André (a,b,c,d), A. Weixlbaumer (a,b,c,d), Mol. Cell 82(20), 3885-3900 (2022); https://doi.org/10.1016/j.molcel.2022.09.020
(a) Department of Integrated Structural Biology, Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Illkirch (France)
(b) Université de Strasbourg, IGBMC UMR 7104 UMR-S 1258, Illkirch (France)
(c) CNRS, UMR 7104, Illkirch (France)
(d) Inserm, UMR-S 1258, Illkirch (France)
| About the beamline: CM01 |
|
Beamline CM01 hosts the ESRF’s Titan Krios cryo-electron microscope (cryo-EM) for single particle experiments. It is equipped with a K3 direct electron detector, a Quantum LS imaging filter and a Volta phase plate. At the ESRF, Cryo-EM is used as a complementary technique to macromolecular crystallography and BioSAXS. Cryo-EM can be used for protein structure determination with near atomic resolution by single-particle imaging to obtain subnanometre-resolution structures of up to ~2 Å including protein complexes and viruses with sizes typically of 150 kDa and above. It is commonly used for larger proteins and complexes that are intrinsically difficult to crystallise. Cryo-EM image analysis is powerful enough to enable in silico classification of different conformation states of the sample if they co-exist in solution. |
Top image: Cryo-EM reconstruction of RNAP (yellow, blue and light grey) bound to a regulatory RNA (red) that promotes its own synthesis.