- Home
- News
- Spotlight on Science
- How archaea connect...
How archaea connect a conserved signal transduction pathway to their rotary motor
11-07-2022
Combining X-ray crystallography data from beamline ID30B and biochemistry with cell biology reveals how the conserved signal transduction machinery in bacteria and archaea is coupled to the archaeal rotary motor termed the archaellum.
Motility is a key feature of many microorganisms and enables them to respond rapidly to changing environmental conditions. Both bacteria and archaea use chemosensory arrays and downstream signal transduction pathways to receive and process external stimuli. These signals are transmitted to a rotary motor that propels the cell forward but can also switch rotational direction leading to a tumbling behaviour and reorientation of the cell [1,2]. While the chemosensory system is conserved among prokaryotes, the function and composition of flagellum and archaellum as major motor structures of bacteria and archaea, respectively, are fundamentally different.
The chemotaxis system in prokaryotes is composed of membrane-embedded receptor proteins that, upon signal reception, trigger a cascade resulting in the phosphorylation of a protein termed CheY. The phosphorylation of CheY leads to small conformational changes within the protein that increase its affinity towards a switch complex on the cytoplasmic face of the flagellum. This interaction of phosphorylated CheY (CheY-P) with the switch complex ultimately leads to a change in the rotational direction of the flagellum. Previous work revealed the molecular details of rotational switching in bacteria (reviewed in [3]). A further study demonstrated that the mechanism of CheY phosphorylation is conserved among bacteria and archaea [4]. However, insights into the molecular details of CheY-P binding to the archaellum and the mechanism of how rotational switching is achieved in the archaellum were still critically lacking.
In contrast to the flagellum, where CheY-P directly binds to the switch complex at the cytoplasmic face, an adaptor protein termed CheF has been identified to be present in all motile archaea [5]. In this study, the crystal structure of full-length CheF was determined using crystallographic data [6] and an initial model obtained from CheF crystals with a strong translational noncrystallographic symmetry.
Click figure to enlarge
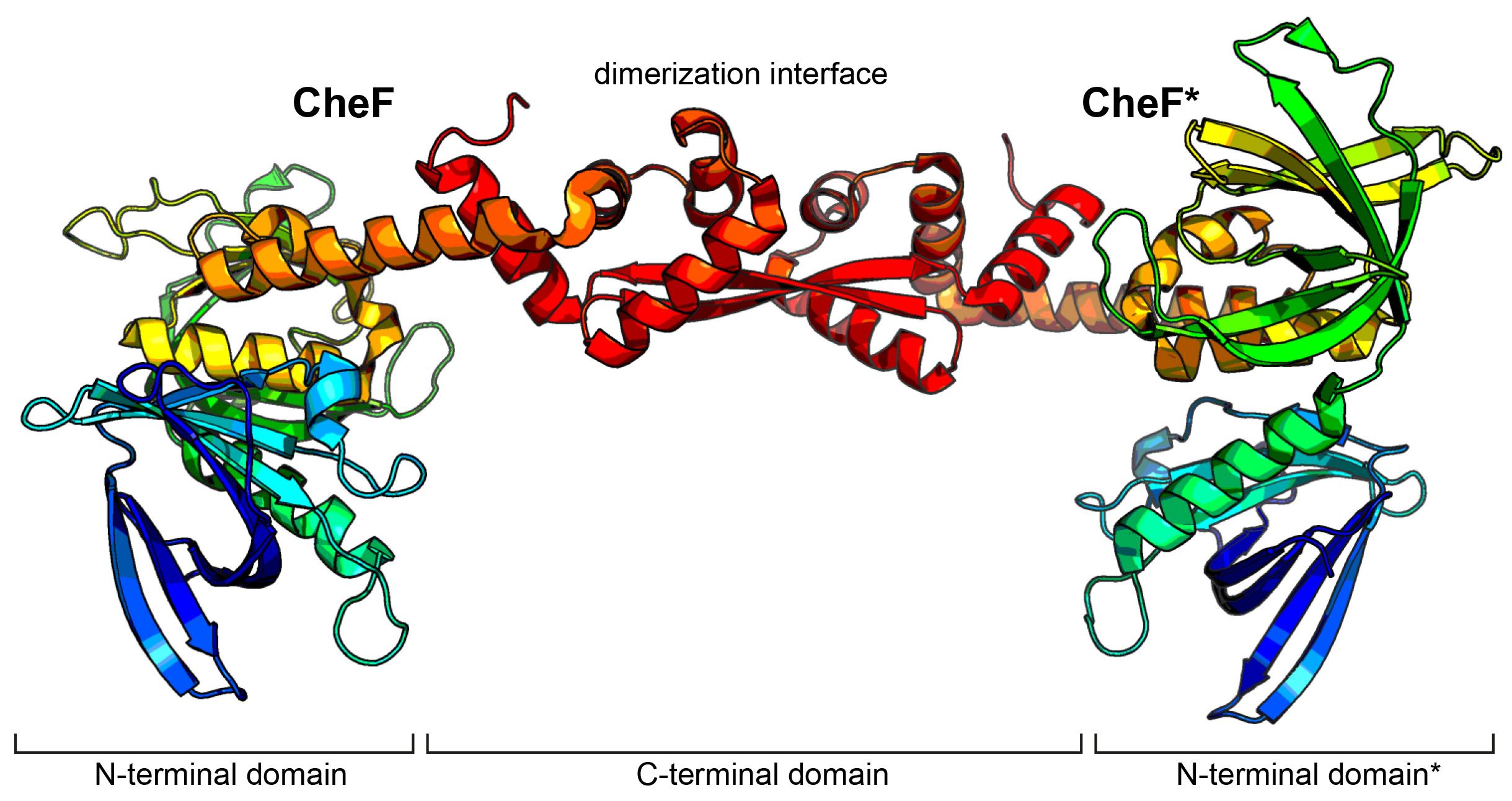
Fig. 1: Crystal structure of CheF. CheF forms an elongated twisted homodimer. The structure has been coloured in rainbow colours from N- to C-terminus.
Figure 1 shows that CheF forms an elongated twisted homodimer consisting of two N-terminal pleckstrin homology domains and a long C-terminal domain (CTD) that also serves as dimerisation interface. This structure made it possible to map and characterise the binding interface between CheF and CheY-P and advised the construction of a fusion protein of the CTD of CheF and CheY. Crystallographic data collected at beamline ID30B made it possible to solve the structure of the CheF-CTD:CheY complex at 2.4 Å resolution. The structural model revealed that two molecules of CheY-P bind to one CheF dimer (Figure 2). Interestingly, the C-terminal carboxyl-moiety of CheF reaches into the phosphorylation site of CheY and therefore might be directly involved in the dephosphorylation of CheY-P. Rapid dephosphorylation of CheY-P is essential to ensure a fast response and avoid having the rotary motor locked in one rotational direction.
Click image to enlarge
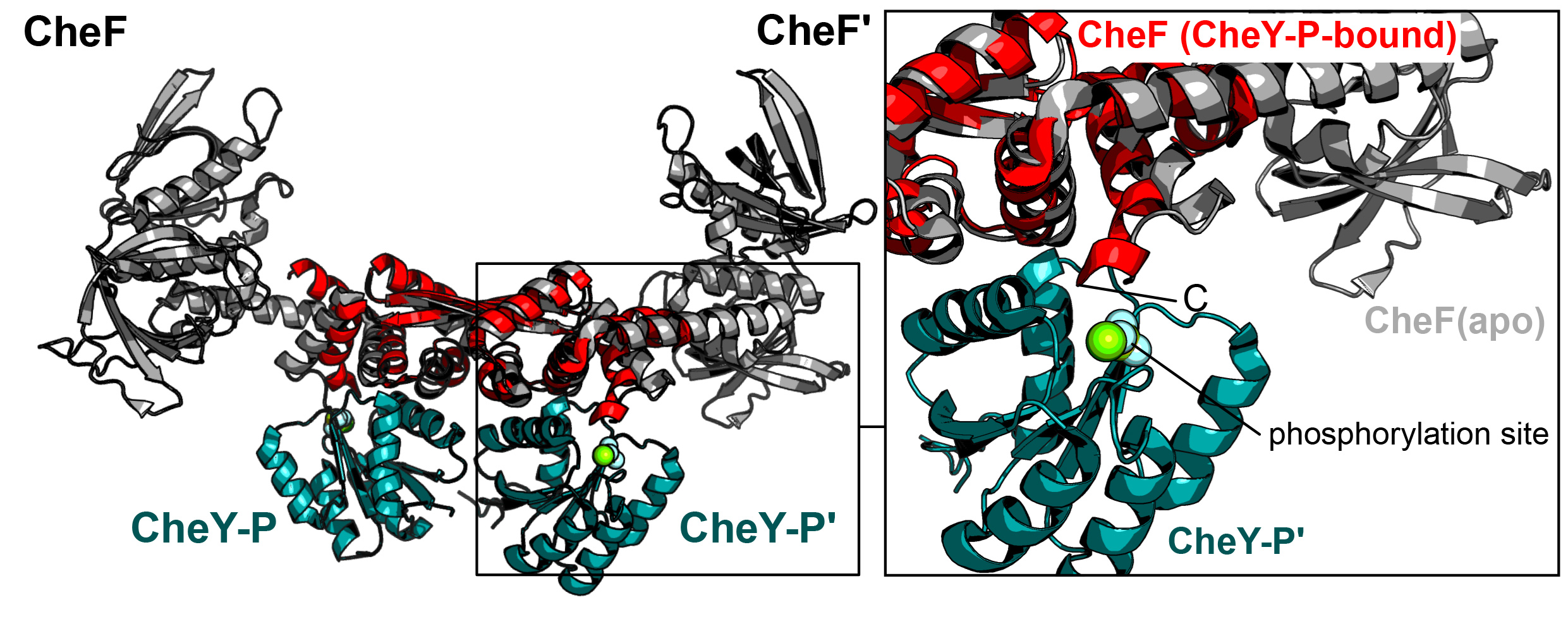
Fig. 2: CheY-P binding induces conformational changes at CheF. Left: Structure of apo CheF (grey) superposed with the CheF-CTD (red) bound to CheY-P (green). Right: Close-up of the interaction interface between CheF and CheY-P showing that the C-terminus of CheF reaches into the phosphorylation site at CheY. Slight conformational changes between the apo and CheY-P-bound states of CheF can be observed.
Furthermore, small conformational changes were observed within CheF in the CheY-P bound structure (Figure 2). It is speculated that the elongated ‘lever-like’ architecture of the CheF dimer might result in even larger conformational changes within the N-terminal domains of CheF that then initiate the rotational switch. However, with the structure of the archaeal switch complex still unknown, future studies need to unveil this connection and the implications of CheY-P binding. Taken together, this study delivers the mechanistic basis of how CheY-P binds to the adaptor protein CheF and suggests a model of how rotational switching of the archaellum might be achieved.
Principal publication and authors
Structural insights into the mechanism of archaellar rotational switching, F. Altegoer #(a,b), T.E.F. Quax (c,d), P. Weiland (a), P. Nußbaum (c), P.I. Giammarinaro (a), M. Patro (c), L. Zhengqun (c), D. Oesterhelt (e), M. Grininger (f), S.V. Albers (c), G. Bange #(a,g), Nat. Commun., 13(1), 2857 (2022); https://doi.org/10.1038/s41467-022-30358-9
(a) Philipps-University Marburg, Center for Synthetic Microbiology (SYNMIKRO) & Faculty of Chemistry, Marburg (Germany)
(b) Current address: Heinrich-Heine University Düsseldorf, Institute of Microbiology, Düsseldorf (Germany)
(c) Archaeal virus-host interactions, Faculty for Biology, University of Freiburg (Germany)
(d) Biology of Archaea and Viruses, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen (The Netherlands)
(e) Max-Planck Institute for Biochemistry, Martinsried (Germany)
(f) Goethe University Frankfurt, Institute of Organic Chemistry and Chemical Biology and
Buchmann Institute for Molecular Life Sciences, Frankfurt am Main (Germany)
(g) Max-Planck Institute for terrestrial Microbiology, Marburg (Germany)
# shared corresponding authors
References
[1] S.V. Albers, K.F. Jarrell, Front Microbiol. pp23 (2015).
[2] H.C. Berg, Annu. Rev. Biochem. 72, 19-54 (2003).
[3] Y. Chang et al., Trends Microbiol. 29, 1024-1033 (2021).
[4] T.E.F. Quax et al., Proc. Natl. Acad. Sci. 115, E1259-E1268 (2018).
[5] M. Schlesner et al., BMC Microbiol. 9(1), 56 (2009).
[6] K. Paithankar et al., Acta Crystallogr. F Struct. Biol. Commun. 75, 576-585 (2019).
|
About the beamline: ID30B ID30B is a highly automated, undulator-based tunable beamline dedicated to macromolecular crystallography, and is the second branch of the canted ID30 beamline. This beamline is tunable between 6 and 20 keV (1.9 – 0.62 Å). Post-EBS, a 2.4 m U35 undulator ensures a high intensity (~1 x 1013 phs/s-1/mm2) X-ray beam. Vertical focusing is achieved at different energies by using CRL combinations. Horizontal focusing is performed by an eliptical mirror in the experimental hutch and contains three polished stripes for harmonic rejection at low, medium and high energies. The beam size at the sample position can be changed between 20 and 200 µm in diameter and is cleaned using beam definition apertures. The experimental setup is currently composed of a MD2-S micro-diffractometer, a new generation FlexHCD sample changer capable of accomodating SPINE standard pins stored in SPINE or Unipuck formats, and a Pilatus3 6M detector. A newly purchased Eiger2X 9M detector is currently being commissioned and should be available for users in late summer 2022. |