- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2011
- Structural biology
- TBL1, a tetrameric scaffold at the heart of the SMRT/NCoR transcriptional repression machinery
TBL1, a tetrameric scaffold at the heart of the SMRT/NCoR transcriptional repression machinery
Transcriptional repression complexes play important roles in both repressing gene transcription and in “resetting” genes after rounds of active transcription.
SMRT and NCoR are homologous corepressor proteins that are recruited to many repressive transcription factors. They are large “platform” or “hub” proteins (c.2500 amino acids) that assemble a complex containing a number of histone deacetylase enzymes. These enzymes remove acetyl groups from histone tails causing chromatin condensation.
The conserved “core” of the SMRT/NCoR repression complexes (called repression domain 1 or RD1) recruits HDAC3, TBL1 (and/or its homologue TBLR1) as well as a protein called GPS2 (also known as AMF1). SMRT, GPS2 and TBL1 form a ternary complex in a three-way interaction.
In this study we sought to understand the mode of assembly of SMRT with TBL1 and GPS2. We mapped the regions in the three proteins required for interaction and demonstrated that the amino-terminal domain of TBL1 (TBL1-NTD) is able to bind both GPS2 and SMRT forming a ternary complex. Adjacent interaction regions in SMRT and GPS2 form a binary complex.
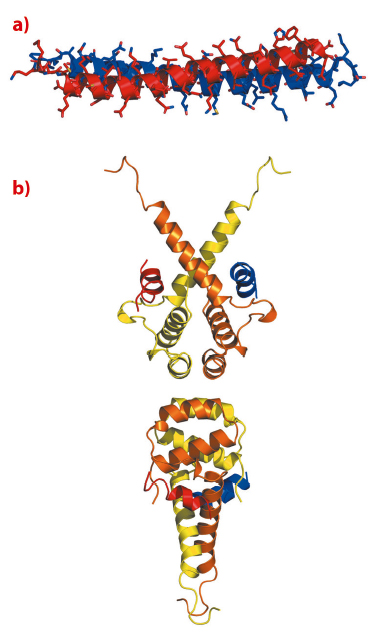
We were unable to obtain crystals of the SMRT:TBL1:GPS2 ternary complex. However, using NMR spectroscopy we were able to determine the structure of the binary complex between SMRT and GPS2. The structure revealed that the two proteins interact through a tight antiparallel coiled-coil (Figure 102a). The antiparallel nature of the coiled-coil positions the regions that interact with TBL1 at the same end of the complex suitable for simultaneous interaction with TBL1 immediately suggesting an arrangement for the ternary complex. The heterodimerisation of SMRT and GPS2 allows them to form a higher affinity complex with TBL1 than could be formed with either alone.
 |
|
Fig. 102: a) NMR structure of the anti-parallel coiled-coil interaction between GPS2 (red) and SMRT (blue). b) Cartoon representation of the crystal structure of tetrameric oligomerisation domain of TBL1 (yellow and orange) with modelled helices binding from GPS2 (red) and SMRT (blue). |
The amino terminal domain of TBL1, that mediates interaction with both SMRT and GPS2, crystallised in three different forms that diffracted to between 2.2 Å and 5 Å resolution.
Unexpectedly this domain formed a tetramer (a dimer of dimers) in all three crystal lattices, with a number of non-polar amino acids interdigitating at the interface between two TBL1 dimers (Figure 102b). Tetramerisation in solution was confirmed by comparing the mobility of wild-type protein and protein with a mutation at the tetramer interface on a size exclusion column. Thus it seems very likely that the tetramer is the physiologically relevant form of TBL1 in vivo. We also showed that each dimer of TBL1 is able to bind a heterodimer of SMRT and GPS2 and thus the physiological relevant stoichiomentry of the TBL1:SMRT:GPS2:HDAC3 complex is 4:2:2:2.
A striking feature of the structure of the TBL1-NTD is that each TBL1 dimer has a pair of non-polar grooves on either side of the structure. These grooves were suggestive of an interaction surface with the SMRT:GPS2 heterodimer. Indeed mutations in the grooves abolished interaction with both SMRT and GPS2. Examination of the GPS2 and SMRT sequences adjacent to the coiled-coil identified a common a-helical sequence motif that could mediate interaction with TBL1. The motif is A x x φ H K/R x φ (where φ is a non-polar amino acid). Mutations in this motif in both proteins abolished recruitment to TBL1. Docking an alpha helix with this motif (using HADDOCK software) indicated a unique mode of binding to the non-polar groove in TBL1 with the key residues making important interactions at the interface.
 |
|
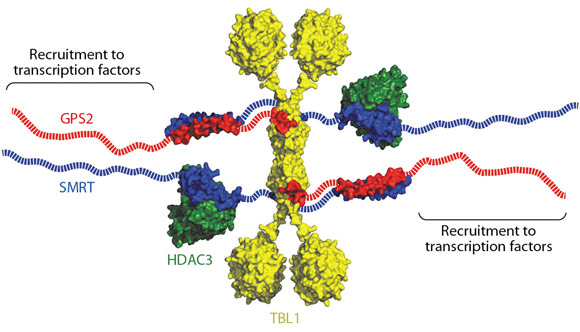
Fig. 103: Schematic of the overall arrangement of the SMRT repression complex, a multivalent chromatin deacetylating machine. |
Taken together, the crystal structure of the TBL1-NTD tetramer, the NMR structure of the SMRT:GPS2 heterodimer, along with mutagenesis and docking experiments give us considerable insight into the assembly of the core SMRT/NCoR:TBL1:GPS2:HDAC3 repression complex. A schematic is shown in Figure 103. In addition to the amino-terminal tetramerisation domain, TBL1 contains a C-terminal WD40 domain that could potentially bind to histone tails and to intact nucleosomes, since such domains have been observed in this role in other proteins. Hence the tetrameric nature of the TBL1 suggests that the protein may serve as a scaffold for a multivalent chromatin-targeted repression machine that contains multiple co-repressor proteins and histone deacetylase enzymes.
Principal publication and authors
J. Oberoi (a,b), L. Fairall (a), P. Watson (a), J-C. Yang (b), Z. Czimmerer (c), T. Kampmann (a), B. Goult (a), J.A. Greenwood (a), J. Gooch (b), B.C. Kallenberger (b), L. Nagy (c), D. Neuhaus (b) and J.W.R. Schwabe (a), Nature Structural and Molecular Biology 18, 177-85 (2011).
(a) Henry Wellcome Laboratories of Structural Biology, Department of Biochemistry, University of Leicester (UK)
(b) Medical Research Council-Laboratory of Molecular Biology, Cambridge (UK).
(c) Apoptosis and Genomics Research Group of the Hungarian Academy of Sciences, Department of Biochemistry and Molecular Biology, University of Debrecen (Hungary).
References
[1] P.J. Watson, L. Fairall and J.W.R. Schwabe, Mol Cell Endocrinol 348, 440–449 (2012).
[2] A. Codina, J.D. Love, Y. Li, M.A. Lazar, D. Neuhaus and J.W.R. Schwabe, Proc Natl Acad Sci USA 102, 6009-6014 (2005).
Acknowledgements
The Wellcome Trust and UK Medical Research Council for funding. Beamlines used for data collection: Beamline ID14-2 at the ESRF; IO3 & IO4 at Diamond Light Source, UK.



