- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2001
- Life Sciences
- The Structure of Bacteriophage T7 Endonuclease I: A Holliday Junction Resolving Enzyme
The Structure of Bacteriophage T7 Endonuclease I: A Holliday Junction Resolving Enzyme
The rearrangement, or recombination, of DNA is an ancient and fundamental biological process. Recombination is central to many diverse biological processes such as the generation of genetic variation (and therefore evolution) and the incorporation of viral DNA into host DNA, resulting in successful viral infection.
The process of DNA recombination occurs in distinct stages (Figure 13), with the formation of a four-way (Holliday) junction as a pivotal intermediate. The penultimate step in DNA recombination is regulated by a junction resolving enzyme or 'resolvase'. This enzyme cleaves the Holliday junction resulting in rearranged DNA strands. Bacteriophage T7 encodes a protein, endonuclease I, which has been shown to act as a four-way DNA junction resolvase.
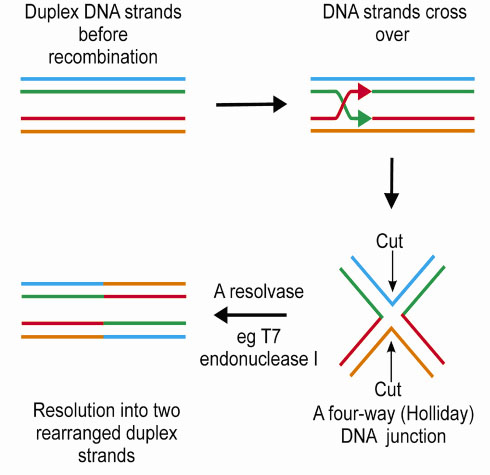 |
Fig. 13: The process of recombination. |
In order to understand the mechanism by which a four-way DNA junction resolving enzyme cleaves DNA we have solved the structure of an inactive mutant of bacteriophage T7 endonuclease I (E65K), using X-ray crystallography. Extensive crystallisation trials showed that high quality protein crystals could only be obtained when the first 11 N-terminal amino acids were removed from the protein. Crystals of endonuclease I (N11, E65K) diffracted X-rays to 2.1 Å on station ID14-EH3.
The crystal structure was solved by the MAD method using data collected from a selenomethionine-substituted protein. However, endonuclease I (N11, E65K) does not contain any endogenous methionine residues. For this reason a methionione containing mutant (I92M) was generated to allow selenomethionine incorporation (endonuclease I,
N11, E65K, I92M). The junction cleavage activity of the protein is unaffected by the introduction of this methionine residue. Selenomethionine-substituted crystals diffracted X-rays rather more weakly than those of the native protein, and three data-sets were collected to 3.0 Å and one to 2.5 Å using station ID14-EH4. All four selenium sites in the asymmetric unit were identified by the SOLVE package, and electron density maps were calculated using the CCP4 suite of programs. The SOLVE derived phases were clear enough to identify protein solvent boundaries and several secondary structure elements, but were significantly improved by density modification and non-crystallographic symmetry averaging. Models were initially built at 2.5 Å using data from the selenomethionine-substituted (
N11, E65K, I92MSe) protein. Later in refinement, 2.1 Å data collected from a crystal of endonuclease I (
N11, E65K) were introduced.
The structure shows that endonuclease I forms a symmetric homodimer arranged in two well-separated domains (Figure 14) and each domain is comprised of elements from both subunits in the dimer. An individual domain comprises a central five-stranded mixed ß-sheet, flanked by five -helices, with one strand and one helix contributed by the other subunit.
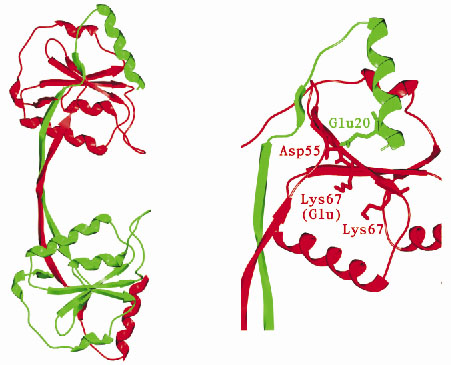 |
Fig. 14: (left) Overall structure of Endonuclease I. (right) Potential active site residues. |
Mutagenesis experiments have previously identified a number of potential active site amino acid residues. Examination of the 3-dimensional arrangement of these residues reveals a close similarity to residues found in the active sites of a number of well-characterised restriction endonucleases. In view of this it is likely that endonuclease I cleaves DNA using a mechanism similar to that of the type II restriction endonucleases. How endonuclease I recognises and cleaves the Holliday junction, while showing no reactivity towards duplex DNA remains an intriguing mystery. The structure of endonuclease I represents a step towards understanding this process.
Principal Publication and Authors
J.M. Hadden (a), M.A. Convery (a, b), A.-C. Déclais (c), D.M.J. Lilley (c) and S.E.V. Phillips (a), Nat. Struct. Biol. 8, 62-67 (2001).
(a) Astbury Centre for Structural Molecular Biology, University of Leeds (UK)
(b) Glaxo SmithKline (UK)
(c) CRC Nucleic Acid Structure Research Group, University of Dundee (UK)



