- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2001
- Life Sciences
- Atomic Model of Cyanobacterial Photosystem I: a Well-defined Assembly of 12 Proteins and 127 Cofactors
Atomic Model of Cyanobacterial Photosystem I: a Well-defined Assembly of 12 Proteins and 127 Cofactors
The conversion of solar energy to chemical energy by photosynthesis forms the main energy source for life on earth. In plants, green algae and cyanobacteria, capable of oxygenic photosynthesis, the central photosynthetic processes of light-induced charge separation are catalysed by two large protein complexes, the photosystems I (PSI) and II (PSII), which are located in the photosynthetic thylakoid membrane.
This year, the crystal structures of both PSI and PSII, isolated from the thermophilic cyanobacterium Synechococcus elongatus, have been published at resolutions of 2.5 Å and 3.8 Å [1], respectively. Until recently, structural studies on PSI by crystallographic methods were limited to a relatively low resolution of 4.0 Å. Significant improvement of the crystal quality, mainly achieved by applying seeding techniques [2], and general improvements in technologies applied in collection of X-ray diffraction data, in particular cryocrystallography, and in crystallographic computing resulted in the present structural model at 2.5 Å resolution. All X-ray data collection was performed at the high-brilliance beamline ID2B at the ESRF. The initial electron density map of PSI was obtained by multiple isomorphous replacement including effects from anomalous dispersion (MIRAS) and permitted the modelling of the 12 protein subunits of PSI.
In cyanobacteria, three monomers are organised into a trimer (Figure 8). In the monomers, the organisation of the subunits in the thylakoid membrane is dictated by the 83 kDa subunits PsaA and PsaB. They form a heterodimer in which the two subunits are related by a pseudo-twofold rotation axis (pseudo-C2). This heterodimer is surrounded by seven smaller membrane intrinsic subunits and three extrinsic subunits located on the stromal side.
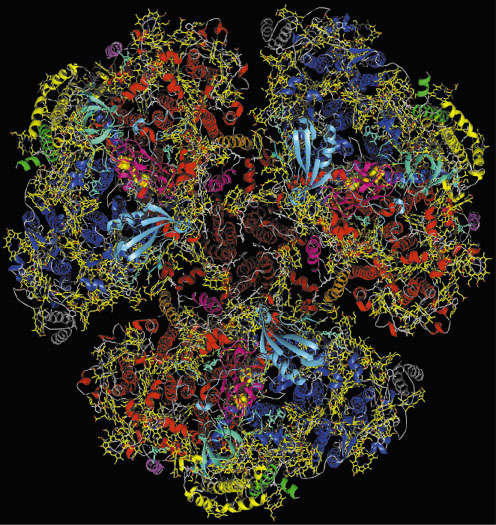 |
Fig. 8: Complete view of cyanobacterial, trimeric photosystem I from the stromal side of the membrane. The 12 protein subunits are in different colours, chlorophylls in yellow, carotenoids in white. |
The charge separation across the membrane is performed by a set of cofactors termed the electron transfer chain (Figure 9). It consists of three pairs of chlorophylls and one pair of phylloquinones (Vitamin K1), positioned along the pseudo-C2 axis in two branches, followed by the 4Fe4S cluster FX that is located right on the pseudo-C2 axis and coordinated to both PsaA and PsaB. The electron is further transferred to the two terminal 4Fe4S clusters FA and FBB located in the stromal subunit PsaC. The light energy required to drive this process is captured by the integral antenna system containing 90 chlorophyll a and 22 carotenoids (Figure 9). The crystal structure of PSI shows new types of Mg2+ axial ligands, not previously observed in the structures of other (bacterio)chlorophyll-protein complexes, the most striking being a phospholipid oxygen and methionine sulphur.
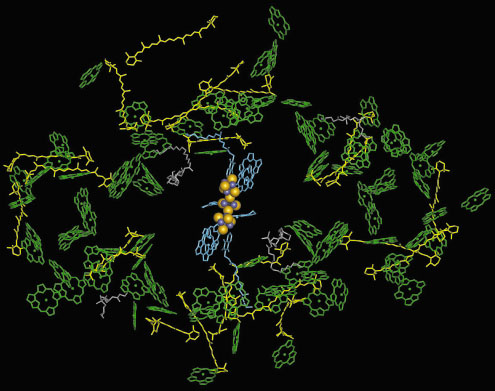 |
Fig. 9: Cofactors of the antenna system (chlorophylls green, ring substituents omitted for clarity, carotenoids yellow) and of the electron transfer chain (blue and Fe4S4 cluster as yellow and blue spheres) in one monomer of photosystem I. |
The structure of PSI provides a detailed picture of the architecture of this protein-cofactor complex. Since the locations and orientations of all the cofactors are known, and their chemical environments are visible for the first time, it is now possible to carry out theoretical studies to understand the spectroscopically determined electron and exciton transfer kinetics between the different cofactors. However, an unambiguous assignment of individual spectral and redox properties of the cofactors is not possible at the moment. The new structural data will initiate mutational studies on PSI which will help to unravel structure-function relationships in more detail.
References
[1] A. Zouni, H.T. Witt, J. Kern, P. Fromme, N. Krauß, W. Saenger and P. Orth, Nature, 409, 739-743 (2001).
[2] P. Fromme and H.T. Witt, Biochim. Biophys, Acta, 1365, 175-184 (1998).
Principal Publication and Authors
P. Jordan (a), P. Fromme (b), H.T. Witt (b), O. Klukas (a), W. Saenger (a) and N. Krauß (a*), Nature, 411, 909-917 (2001).
(a) Institut für Chemie/Kristallographie, Freie Universität Berlin (Germany)
(b) Institut für Chemie, Max-Volmer-Laboratorium, Technische Universität Berlin (Germany)
* present address: Institut für Biochemie, Universitätsklinikum Charité, Humboldt-Universität zu Berlin (Germany)



