- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2001
- Life Sciences
- Extending the Range of the de novo Phasing of Native Protein Crystal Structures using the Anomalous Scattering from Sulphur Atoms
Extending the Range of the de novo Phasing of Native Protein Crystal Structures using the Anomalous Scattering from Sulphur Atoms
The macromolecular crystallography community is increasingly interested in new, routine methods of structure solution which require neither the chemical modification of macromolecules nor the introduction of very heavy atoms into crystals. One technique that has recently received a great deal of attention is the exploitation, using X-rays of relatively long wavelength ( > 1.5 Å), of the significant anomalous scattering properties of sulphur atoms found in the structures of almost all proteins [1, 2]. For all of the macromolecular crystal structures thus far solved using this method, the crystals diffracted to much better than 2 Å resolution. The method would thus appear to require crystals that diffract rather well. It is also unclear just what the limits are for the size of anomalous signal that can successfully be used in this procedure.
In order to ascertain just how general a method S-SAD (Sulphur-Single-Wavelength Anomalous Dispersion) can become and what the limits of the technique are, we have solved the structures of two crystal forms of Tryparedoxin II using data collected on BM14 at 1.77 Å wavelength. The two crystal forms diffract to1.5 Å and 2.7 Å resolution respectively and both experiments produced extremely high quality, interpretable electron density maps (see Figure 6).
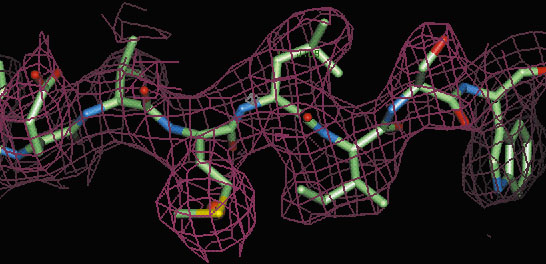 |
| Fig. 6: Part of the electron density map (red chicken wire) for crystal form II of Tryparedoxin II calculated using solvent flattened S-SAD phases at 2.7 Å resolution. |
The success of the latter experiment extends the resolution limits of the technique markedly, shows that the technique can be successful even using data from crystals that diffract only to medium resolution and has significant implications for macromolecular structure determination in the high throughput era. In the 10 years to mid-October 2001, the number of macromolecular crystal structures submitted to the Protein Data Bank was 12633. Of these 11679, or 92.5%, were determined to a resolution of 2.7 Å or better. Thus, even allowing for the skewing effect of amino acid mutant structures etc., the vast majority of crystal structures would, in terms of resolution of data available, be amenable to solution by S-SAD. However, one must also take into account amino acid composition when considering whether a protein crystal structure might be suitable for solution by S-SAD. Of the ordered amino acid residues in the asymmetric unit of the crystals studied here, those containing sulphur represent 4.7%. This produces, at = 1.77Å, an anomalous signal from crystals of the native protein that is strong enough to allow structure solution of both crystal forms. Of the five eukaryotic genomes for which there is full or partial sequence information, the frequency of occurrence of sulphur-containing amino acid residues is as follows (see http://www.ebi.ac.uk/proteome/ for details): Homo sapiens, 4.4%; Arabidopsis thaliana, 4.3%; Caenorhabditis elegans, 4.7%; Drosophila melanogaster, 4.2%; Saccharomyces cerevisiae, 3.4%. Large numbers of proteins from all of these genomes should thus, in principle, be amenable to structure solution by S-SAD (see Figure 7 for a breakdown of % sulphur-containing residues for C. elegans) and our experiments have therefore shown that S-SAD has the potential to become a major technique for macromolecular crystal structure determination.
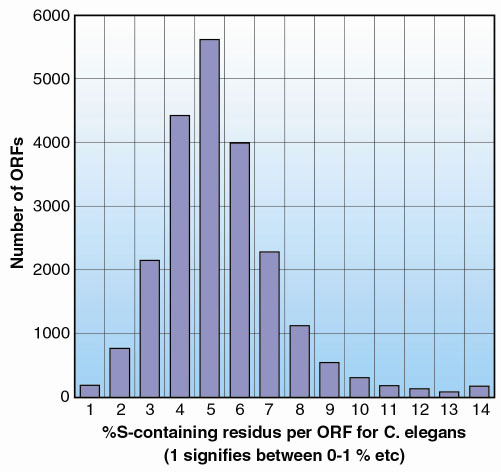 |
Fig. 7: A histogram showing the distribution of the number of open reading frames (ORFs) as a function of the percentage content of sulphur containing residues for the genome of C. elegans. As can be seen more than 50% of all ORFs in this genome are predicted to produce proteins containing at least as high a ratio of sulphur containing residues as found in Tryparedoxin II. |
References
[1] Z. Dauter, M. Dauter, E. de la Fortelle, G. Bricogne and G.M. Sheldrick, J. Mol. Biol., 289, 83-92 (1999).
[2] Z-J. Lui, E.S. Vysotski, C-J. Chen, J.P. Rose, J. Lee and B.C. Wang, Protein Science 9, 2085-2093 (2000).
Principle Publication and Authors
E. Micossi (a), W.N. Hunter (b) and G.A. Leonard (a), Acta Cryst., D58, 21-28 (2001).
(a) ESRF
(b) School of Life Sciences, University of Dundee, Scotland (UK)



