- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2001
- Life Sciences
- Hybrid Cluster Proteins (HCP): A Unique Biological Metal Centre
Hybrid Cluster Proteins (HCP): A Unique Biological Metal Centre
Hybrid cluster proteins (HCP), first reported in 1989 [1], have unprecedented redox chemistry and are unique amongst metal centres in biological systems. Despite the wealth of spectroscopic and structural information on HCP, the precise physiological function(s) of these proteins remains unknown. To account for the early electron paramagnetic resonance analyses carried out on the protein, the presence of a [6Fe-6S] cluster was proposed and the protein was therefore named the "prismane protein" after the shape of the proposed iron-sulphur cluster. The first X-ray crystal structure of the protein, isolated aerobically from Desulfovibrio vulgaris (Dv), showed that HCP does not contain a [6Fe-6S] cluster, but instead has two independent centres each containing four iron atoms [1]. One of these is a cubane [4Fe-4S] structure and the second is the so-called "hybrid cluster", a novel [4Fe-2S-2O] cluster with an unusual arrangement.
The quite unusual and unexpected structure of the hybrid cluster raised the question of whether it was indeed a native structure. The similar protein from Desulfovibrio desulfuricans (Dd) was therefore purified and crystallised anaerobically. X-ray data for the Dd protein were collected at a wavelength of 1.722 Å on BM14, so that anomalous dispersion effects could be used to confirm the locations of the iron atoms in the clusters. Then a second high-resolution data set, 1.25 Å, was collected using a wavelength of 0.933 Å on ID14-2.
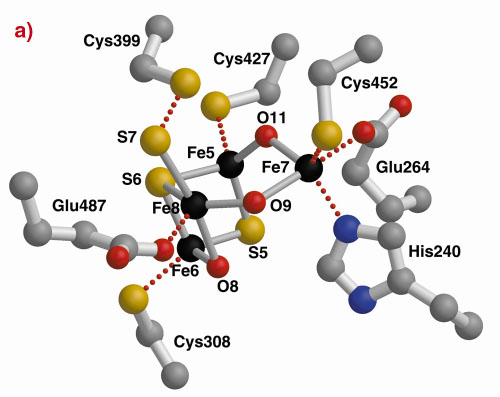
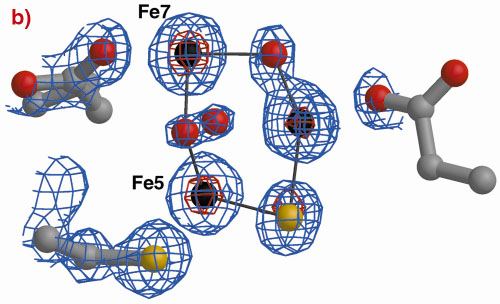
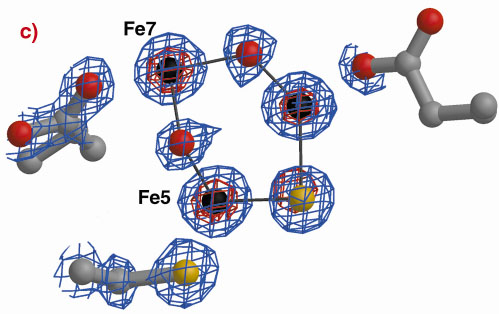
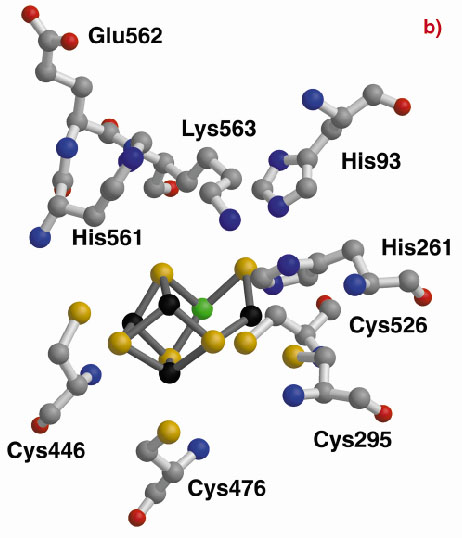
The three-dimensional structures of the Hybrid Cluster Proteins from Dd and Dv were shown to have a very high similarity and both contain a cubane and a hybrid cluster. The overall protein and cluster structure appears to be independent of the oxidation state of the protein and/or whether the preparations were performed aerobically or anaerobically. The hybrid clusters contain both oxygen and sulphur bridges between pairs of iron atoms (Figure 3) and a further moiety, X, appears to bridge Fe5 and Fe7 thereby completing their coordination geometries. The hybrid cluster itself has an open configuration and is readily accessible by both hydrophobic cavities and hydrophilic channels [2]. The position of X represents an obvious site of substrate binding and Fe8 may also be involved, but the nature of the substrate and the reaction mechanism both remain to be clarified.
|
|
Fig. 3: The hybrid cluster in the HCP proteins. (a) A schematic view, (b) electron density in a 2|Fo|-|Fc| map contoured at the 2.0 (blue) and 15.0 (red) rms levels in the vicinity of the X moiety in the Dv protein, and (c) as per (b) for molecule A in the Dd protein. |
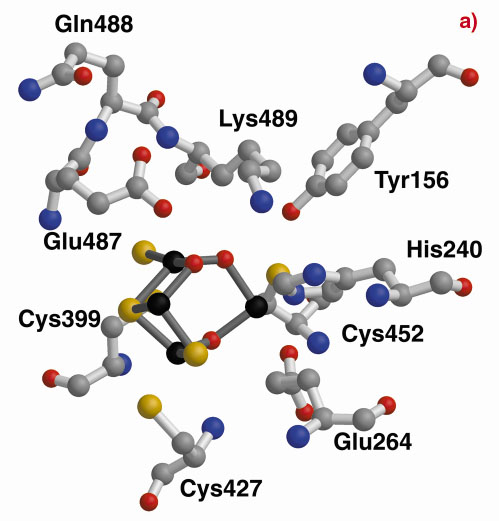
A structurally-based sequence alignment between the HCP and the carbon monoxide dehydrogenase (CODH) enzyme from C. hydrogenoformans highlights the close structural similarity between Dd/Dv and C. hydrogenoformans (Figure 4). In fact, all the Dd/Dv cysteine and histidine cluster binding residues and many of the residues contributing to the strong hydrophobicity of one of the cavities pointing towards Fe8 [2] are conserved. This cavity leads directly from the surface of the protein to Lys489, a residue that is retained between many HCP and CODH and located within 3.0 Å of the oxygen atom bridging Fe7 and Fe8 in the HCP. This lysine is predicted to have a crucial role in the CODH enzyme mechanism [3] and may indicate a hydrophobic pathway for substrate or product with a common key role for Lys489 between the CODH and the HCP reaction mechanisms. However, so far, any attempts to find such activity have failed.
|
|
Fig. 4: The hybrid clusters of the Dd HCP (a) and the CODH-cooS from C. hydrogenoformans (b) after structural alignment. In the vicinity of the hybrid clusters, the cysteine, Cys308 (not shown in the figure), Cys399, Cys427 and Cys452, and histidine, His240, cluster binding residues are conserved. |
More recent results have suggested a possible sulphur transferase role for the HCP proteins and this aspect is now being vigorously pursued.
References
[1] W.R. Hagen, A.J. Pierik and C. Veeger, J. Chem. Soc. Faraday Trans I 85, 4083-4090 (1989).
[2] S.J. Cooper, C.D. Garner, W.R. Hagen, P.F. Lindley and S. Bailey, Biochem., 39, 15044-15054 (2000).
[3] H. Dobbek, V. Svetlitchnyi, L. Gremer, R. Huber and O. Meyer, Science, 293, 1281-1285 (2001).
Principal publication and authors
S. Macedo (a, b), E.P. Mitchell (b), C.V. Romão (a), S.J. Cooper (c, d), R. Coelho (a), M.Y. Liu(e), A.V. Xavier (a), J. LeGall (e), S. Bailey (c*), C.D. Garner (d),W.R. Hagen (f), M. Teixeira (a), M.A. Carrondo (a) and P. Lindley (b), to be published.
(a) Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Oeiras (Portugal)
(b) ESRF
(c) CLRC Daresbury Laboratory, Warrington (UK)
(d) School of Chemistry, University Park, Nottingham (UK)
(e) Department of Biochemistry, University of Georgia (USA)
(f) Delft University of Technology, Kluvyer Department of Biotechnology (The Netherlands)
* Current address: Lawrence Berkeley National Laboratory, Berkeley (USA)