- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2001
- Life Sciences
- The Crystal Structure of an Asymmetric Complex of the Two Nucleotide Binding Components of Proton-Translocating Transhydrogenase
The Crystal Structure of an Asymmetric Complex of the Two Nucleotide Binding Components of Proton-Translocating Transhydrogenase
Membrane-bound ion translocators have important functions in biology, but their mechanisms of action are poorly understood. Transhydrogenase, found in animal mitochondria and bacteria, links the redox reaction between NAD(H) and NADP(H) to proton translocation across a membrane. The enzyme is a dimer in the membrane. Each protomer consists of three domains: dI and dIII protrude from the membrane and bind NAD(H) and NADP(H), respectively; dII spans the membrane and provides a channel for proton translocation. Depending on enzyme source, the protomer may consist of 1, 2 or 3 polypeptide chains.
Under most physiological conditions, transhydrogenase consumes the proton electrochemical gradient (p) generated by the respiratory (or photosynthetic) electron transport chain:
NADH + NADP+ + H+out Æ NAD+ + NADPH + H+in
Thus, the energy of p is utilised to drive the reaction toward NADP+ reduction. NADPH is used subsequently in biosynthesis and to reduce glutathione for detoxification reactions. The transfer of hydride-ion equivalents between NAD(H) and NADP(H) in transhydrogenase is direct, and, therefore, it is envisaged that the nicotinamide rings of the two nucleotides are brought into apposition to effect the redox reaction.
Recombinant dI and dIII from R. rubrum spontaneously form an active complex in solution that can reduce NADP+ by NADH, even though dII, the membrane-spanning domain is absent. Figure 18 shows the structure of a dI2:dIII1 complex from R. rubrum in the presence of both NAD+ and NADP+. Two equivalents of dI form a tight dimer. One dI (subunit A) has tightly-bound NAD+, while subunit B does not bind NAD+ tightly, but instead has a bound dIII containing tightly-bound NADP+. This remarkably asymmetric complex has been shown by a number of techniques to be functionally relevant [1].
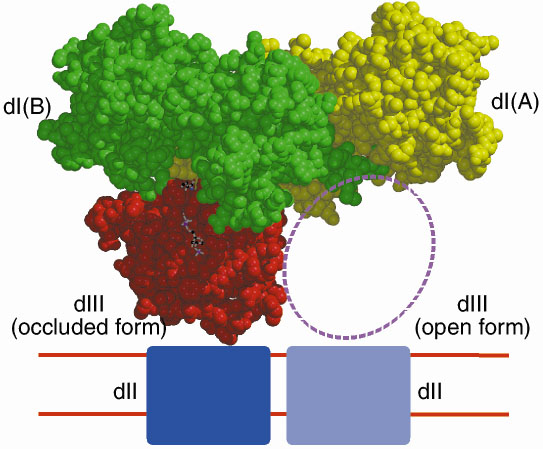 |
| Fig. 18: Structure of a dI2:dIII1 complex from R. rubrum. |
NAD+ from dI subunit A can be easily modelled into the, presumably, partially occupied, or empty, NAD(H)-binding site of subunit B. This shows that although the two nucleotides NADP+ and the modelled NAD+ are close in space, they are too far apart for direct hydride transfer between the two NC4 atoms to occur. However, by simple rotation about a few bonds, the nicotinamide ring of NAD+ can be brought into apposition of NADP+, allowing us to model the hydride-transfer step. Figure 19 shows the dI:dIII interface.
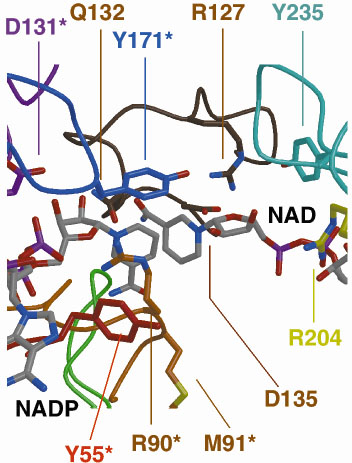 |
Fig. 19: The dI:dIII interface. |
Together with earlier experimental observations, this structure is taken to indicate that proton pumping is achieved through an alternating NADP(H) binding change mechanism.
Reference
[1] J.D. Venning, D.J. Rodrigues, C.J. Weston, N.P.J. Cotton, P.G. Quirk, N. Errington, S. Finet, S.A. White and J.B. Jackson, J. Biol. Chem., 276, 30678-30685 (2001).
Principal Publication and Authors
N.P.J. Cotton (a), S.A. White (a), S.J. Peake (a), S. McSweeney (b) and J.B. Jackson (a), Structure, 9, 165176, (2001).
(a) School of Biosciences, University of Birmingham, Birmingham (UK)
(b) ESRF



